













| General Information: |
|
| Name(s) found: |
TEA1 /
YOR337W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 759 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IGI |
| Biological Process: |
transcription
[IGI |
| Molecular Function: |
DNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAPLWPNKN EKNHTVKRAL STDMTSNILS STNASSNEEN SRSSSAANVR SGTGANTLTN 60
61 GGSTRKRLAC TNCRNRRKKC DLGFPCGNCS RLELVCNVND EDLRKKRYTN KYVKSLESHI 120
121 AQLETNLKNL VQKIYPDDEQ ILNRMMVGDV LSALPDSSQV SINYTDQTPS LPIPATRGTF 180
181 IIENDKVSQP LSSFNQQTEP STLNSGIFNT QKQNFEESLD DQLLLRRSLT PQGEKKKKPL 240
241 VKGSLYPEGP VSYKRKHPVK SDSLLPVSSL TAATDPSTFS DGITAGNSVL VNGELKKRIS 300
301 DLKTTVIVRG LNDDNPNSIN NDPRILKSLS NFYKWLYPGY FIFVHRESFL YGFFNHSKNN 360
361 YEDSSYCSVE LIYAMCAVGS RLTPDLQEYS EVYYQRSKKT LLQLVFDEQS TARITTVQAL 420
421 FCLAFYELGK GNNQLGWYFS GLAIRVGYDM GFQLDPKVWY VDDNNLQLTQ SELEIRSRIY 480
481 WGCYIADHFI CLMLGRTSTL SVSNSTMPES DELPEVNGTE EFRFIGRHVL QISLPLKNLI 540
541 ILSRLVQIFT SKIFIESEDI ARKLKYLNTF NSQVYNWRQS LPEFLQWSKT LIENDDVSTD 600
601 PTISYFWYCY YIVRLTFNKP FIEDSQESET VVIEIIDDLK TLLDNFGKKF GNYTKGNLYQ 660
661 LYSCLLAINC LKKLKEIRSS EQDSWNAQLD FFNHIFYTQL YPAYDLPKKL QEDTELETEQ 720
721 ENQMLNQVGN INYTHDFSLS HEIDDLIREL FGVGTPQKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
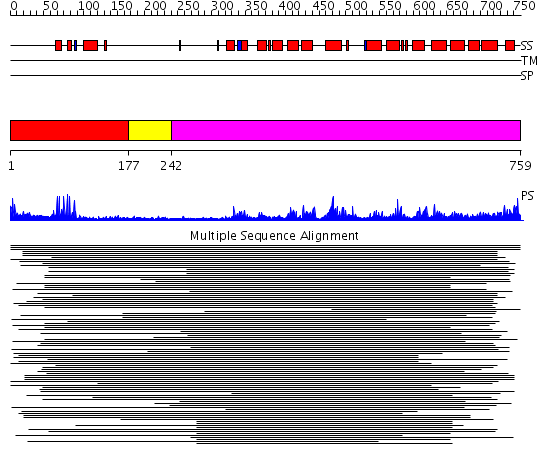
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 34.38764 | PPR1 | |
| 2 | View Details | [177..241] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [242..759] | 15.21099 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [512..759] | 2.337994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)