













| General Information: |
|
| Name(s) found: |
TRM13 /
YOL125W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 476 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
tRNA methylation
[IDA |
| Molecular Function: |
tRNA methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLQDNNGPAV KRAKPSERLQ CEYFMEKKKR RCGMTRSSQN LYCSEHLNLM KKAANSQVHN 60
61 KNGSEAEKER ERVPCPLDPN HTVWADQLKK HLKKCNKTKL SHLNDDKPYY EPGYNGENGL 120
121 LSSSVKIDIT AEHLVQSIEL LYKVFEGESM DELPLRQLNN KLMSLKRFPQ LPSNTKHAVQ 180
181 QSSLIENLVD AGAFERPESL NFIEFGCGRA EFSRYVSLYL LTQLTSLPAE HSGSNSNEFV 240
241 LIDRATNRMK FDKKIKDDFS EIKSNSPSKP ISCPSIKRIK IDIRDLKMDP ILKSTPGDDI 300
301 QYVCISKHLC GVATDLTLRC IGNSSILHGD DNNGCNPKLK AICIAMCCRH VCDYGDYVNR 360
361 SYVTSLVEKY RAHGSILTYE TFFRVLTKLC SWGTCGRKPG TAITDIVNVV ESFEGAEPYT 420
421 ITIKERENIG LMARRVIDEG RLVYVKEKFT EFNAELIRYV ESDVSLENVA MLVYKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
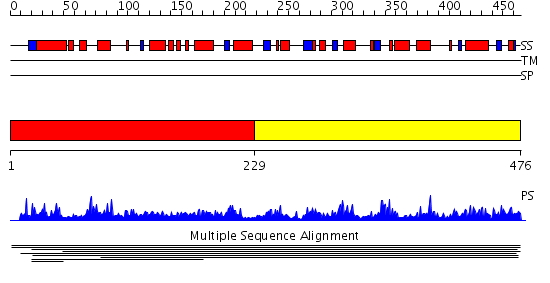
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..228] | 1.007954 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [229..476] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [309..353] | 0.996 | SOLUTION STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR II RELATIONSHIP TO RECEPTOR AND BINDING PROTEIN INTERACTIONS | |
| 4 | View Details | [354..476] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)