













| General Information: |
|
| Name(s) found: |
ABZ1 /
YNR033W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 787 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
para-aminobenzoic acid metabolic process
[IMP |
| Molecular Function: |
4-amino-4-deoxychorismate synthase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSDTIDTKQ QQQQLHVLFI DSYDSFTYNV VRLIEQQTDI SPGVNAVHVT TVHSDTFQSM 60
61 DQLLPLLPLF DAIVVGPGPG NPNNGAQDMG IISELFENAN GKLDEVPILG ICLGFQAMCL 120
121 AQGADVSELN TIKHGQVYEM HLNDAARACG LFSGYPDTFK STRYHSLHVN AEGIDTLLPL 180
181 CTTEDENGIL LMSAQTKNKP WFGVQYHPES CCSELGGLLV SNFLKLSFIN NVKTGRWEKK 240
241 KLNGEFSDIL SRLDRTIDRD PIYKVKEKYP KGEDTTYVKQ FEVSEDPKLT FEICNIIREE 300
301 KFVMSSSVIS ENTGEWSIIA LPNSASQVFT HYGAMKKTTV HYWQDSEISY TLLKKCLDGQ 360
361 DSDLPGSLEV IHEDKSQFWI TLGKFMENKI IDNHREIPFI GGLVGILGYE IGQYIACGRC 420
421 NDDENSLVPD AKLVFINNSI VINHKQGKLY CISLDNTFPV ALEQSLRDSF VRKKNIKQSL 480
481 SWPKYLPEEI DFIITMPDKL DYAKAFKKCQ DYMHKGDSYE MCLTTQTKVV PSAVIEPWRI 540
541 FQTLVQRNPA PFSSFFEFKD IIPRQDETPP VLCFLSTSPE RFLKWDADTC ELRPIKGTVK 600
601 KGPQMNLAKA TRILKTPKEF GENLMILDLI RNDLYELVPD VRVEEFMSVQ EYATVYQLVS 660
661 VVKAHGLTSA SKKTRYSGID VLKHSLPPGS MTGAPKKITV QLLQDKIESK LNKHVNGGAR 720
721 GVYSGVTGYW SVNSNGDWSV NIRCMYSYNG GTSWQLGAGG AITVLSTLDG ELEEMYNKLE 780
781 SNLQIFM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
ABZ1 MPA43 TSA1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
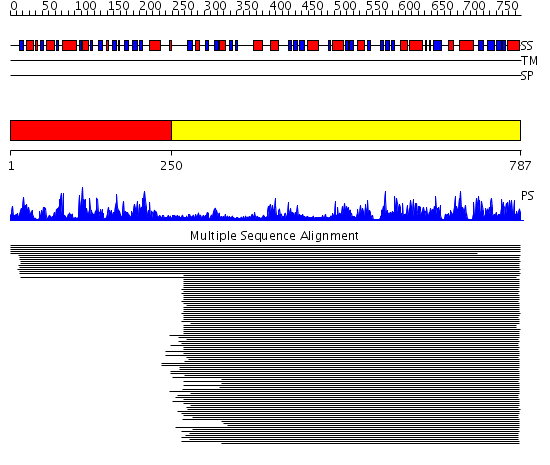
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..249] | 257.855515 | Anthranilate synthase GAT subunit, TrpG | |
| 2 | View Details | [250..787] | 481.457575 | P-aminobenzoate synthase component I |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)