













| General Information: |
|
| Name(s) found: |
VNX1 /
YNL321W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 908 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKNNHISAS GNSTSGDHRL KEEVLTPTTS ASTPHRIFSV DDDPKEIQND IRYLEGLHEG 60
61 LKFALHANKS KRSVSSQSPI VHSSNNTLHH HEHQQHLPPT LESLSSKSHS VPDLNTATPS 120
121 SPKRMHSSIR ELPHDDNDDE DANDDSRFII HDSHGHDLLI DEINCQSPSH LENNDQASNA 180
181 SSTESFTLRE RQDAINETHP FGIRIWKPAL YKKHRSVQRT AAQDIHETQL KTITWEVTCS 240
241 NVLWFILFGF PIAILFYSAA IVVFLLGGGG LVTNSAKEYS KCLYKLANYF LWPFGKMVYL 300
301 LQDEQYLQED KDEGISMQQF YNWVTSYSNR LVFHQSQAKF QQREDHPAPA TESSSLMPPA 360
361 NTTATPLNSN HPSYNSIRHE IPHAAAQRRY FGRGKWSWGR VLFYTIFHLV LQPILAVLSL 420
421 CLWLLVFTIP MSNVLWQIMY HCRRHPLALG FKYVENSSQS HENEITQQQL NKNILLCTFR 480
481 AAGWHYYKYT VDGTNVIVVN LISIVFFTIF DFYVLKNFLH WKTWFTYESS IFILCLTSTI 540
541 PLAFYIGQAV ASISAQTSMG VGAVINAFFS TIVEIFLYCV ALQQKKGLLV EGSMIGSILG 600
601 AVLLLPGLSM CGGALNRKTQ RYNPASAGVS SALLIFSMIV MFVPTVLYEI YGGYSVNCAD 660
661 GANDRDCTFS HPPLKFNRLF THVIQPMSIS CAIVLFCAYI IGLWFTLRTH AKMIWQLPIA 720
721 DPTSTAPEQQ EQNSHDAPNW SRSKSTCILL MSTLLYAIIA EILVSCVDAV LEDIPSLNPK 780
781 FLGLTIFALI PNTTEFLNAI SFAIHGNVAL SMEIGSAYAL QVCLLQIPSL VIYSIFYTWN 840
841 VKKSMINIRT QMFPLVFPRW DIFGAMTSVF MFTYLYAEGK SNYFKGSMLI LLYIIIVVGF 900
901 YFQGALSE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
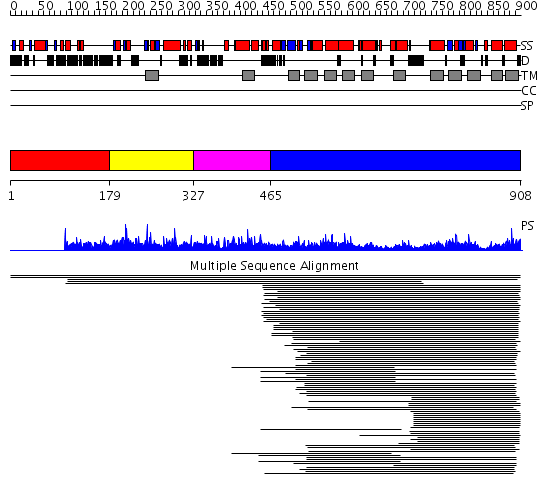
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [179..326] | 32.187087 | Domain of unknown function (DUF307) No confident structure predictions are available. | |
| 3 | View Details | [327..464] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [465..908] | 41.116994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [720..908] | 20.769551 | No description for PF01699.15 was found. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|