













| General Information: |
|
| Name(s) found: |
CHA4 /
YLR098C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 648 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI |
| Biological Process: |
cellular amino acid catabolic process
[IGI regulation of transcription, DNA-dependent [IMP |
| Molecular Function: |
transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMLEPSPPPL TTTVTPSLPS SLKKSVTDND QNNNNVPRKR KLACQNCRRR RRKCNMEKPC 60
61 SNCIKFRTEC VFTQQDLRNK RYSTTYVEAL QSQIRSLKEQ LQILSSSSST IASNALSSLK 120
121 NNSDHGDAPN EKILKYGETA QSALPSSESN DENESDAFTK KMPSESPPPV GTNSIYPSNS 180
181 LSIIKKKTDG STRYQQQQVS LKNLSRSPLI LRSLSLFFKW LYPGHYLFIH RETFLSAFFG 240
241 DTNTKSYYCS EELVFAIAAL GSLISYKSET ELFQQSEVFY QRAKTIVLKK IFQLEDSSLA 300
301 ESSSSSKLAI IQTLLCLAFY DIGSGENPMA WYLSGLAFRI AHEIGLHLNP EAWSNVYEDE 360
361 LSIMDFEVRS RIYWGCYIAD HLIAILFGRS TSLRLSNSTV PETDELPEIE TGIEEYIYDP 420
421 KVILSTANPL KKLIVLSRIT EIFASKIFSP NETLLQRSEY LAKFNLEVYN WRRDLPPELQ 480
481 WTKRSLMEMT DFNPTIAYVW FHYYIVLISY NKPFIYEIKQ SRELVEGYVD ELYYLLKVWK 540
541 NKFKTFEKAT IYMIYSAILA IQCMKSNLIK KDRKQDFLNF LSAPTLNYEL ARKFIENSED 600
601 ALHNSETMDL LGTLSHGNDF ALEYNFDFTL LNEIDMLIGG NTNDGLSK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
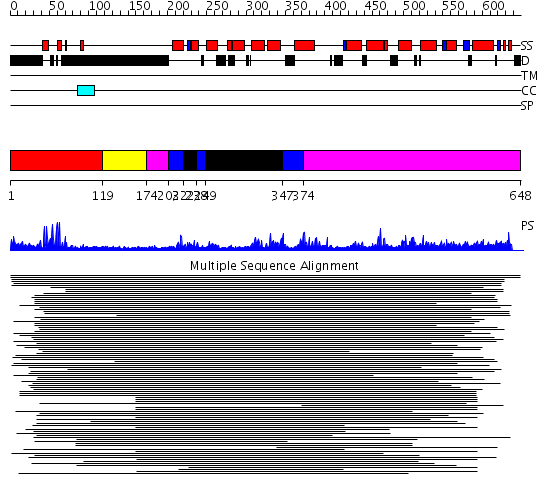
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 36.30103 | CD2-Lac9 | |
| 2 | View Details | [119..173] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [174..202] [374..648] |
7.23 | (+)-bornyl diphosphate synthase | |
| 4 | View Details | [203..221] [238..248] [347..373] |
7.23 | (+)-bornyl diphosphate synthase | |
| 5 | View Details | [222..237] [249..346] |
7.23 | (+)-bornyl diphosphate synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)