













| General Information: |
|
| Name(s) found: |
RTT109 /
YLL002W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 436 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
histone acetylation
[IDA negative regulation of transposition, DNA-mediated [IMP response to DNA damage stimulus [IMP |
| Molecular Function: |
histone acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLNDFLSSV LPVSEQFEYL SLQSIPLETH AVVTPNKDDK RVPKSTIKTQ HFFSLFHQGK 60
61 VFFSLEVYVY VTLWDEADAE RLIFVSKADT NGYCNTRVSV RDITKIILEF ILSIDPNYYL 120
121 QKVKPAIRSY KKISPELISA ASTPARTLRI LARRLKQSGS TVLKEIESPR FQQDLYLSFT 180
181 CPREILTKIC LFTRPASQYL FPDSSKNSKK HILNGEELMK WWGFILDRLL IECFQNDTQA 240
241 KLRIPGEDPA RVRSYLRGMK YPLWQVGDIF TSKENSLAVY NIPLFPDDPK ARFIHQLAEE 300
301 DRLLKVSLSS FWIELQERQE FKLSVTSSVM GISGYSLATP SLFPSSADVI VPKSRKQFRA 360
361 IKKYITGEEY DTEEGAIEAF TNIRDFLLLR MATNLQSLTG KREHRERNQP VPASNINTLA 420
421 ITMLKPRKKA KALPKT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
RTT109 VPS75 |
| View Details | Krogan NJ, et al. (2006) |
|
FMS1 FUS3 RTT109 VPS75 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Huh WK, et al. (2003) |
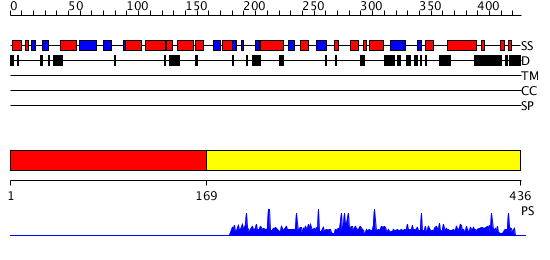
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [169..436] | 1.0029 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)