













| General Information: |
|
| Name(s) found: |
STR2 /
YJR130C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 639 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
sulfur metabolic process
[IMP |
| Molecular Function: |
cystathionine gamma-synthase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISRTIGESI PPNTKHAVSV CLPTWEATVG YEEGESSIIN SLTTGYPRFF IHKSIKKLCE 60
61 ILSAKYSMED EACLCFPSYK VANRCREFIK VKTGLSTKVR ILQLCTPKPM NQEEKLWRRE 120
121 CKITVVFVDQ EIFPVMKQYW QHSGEIVSSR MAEYILHELQ VKDNLKKMET VDNGKKFMTE 180
181 DENRVNEEYI ETRFGRNLNF LAADKAKYLI RKRIATKVVE KIDSEGLSDL FSFEHYNESN 240
241 GPFNVGSGEA LDDDQLNSDI PAETITSMGE SGSNSTFENT ATDDLKFHVN PNTDVYLFPS 300
301 GMASIFTAHR LLLNFDAKRL SRSSSRQDKL IGYGPPFKKT VMFGFPYTDT LSILRKFNHT 360
361 HFLGQGDSTS MNALKNILHS GEQILAVFIE APSNPLLKMG DLQELKRLSD LYSFYIVVDE 420
421 TVGGFVNIDV LPYADIVCSS LTKIFSGDSN VIAGSLVLNP RGKIYEFARK FMKTEDGYED 480
481 CLWCEDALCL ERNSRDFVER TIKVNTNTDI LLKRVLLPQV GKLFKKIYYP SLTSEDTKRN 540
541 YDSVMSTKDG GYGGLFSLTF FNIEEAKKFF NNLELCKGPS LGTNFTLACP YAIIAHYQEL 600
601 DEVAQYGVET NLVRVSVGLE NSDVLCNVFQ RAIEKALGE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
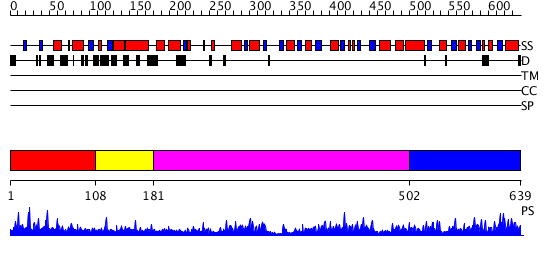
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 2.221849 | Prepeptide of 5-ALAS | |
| 2 | View Details | [108..180] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [181..501] | 208.457575 | Methionine gamma-lyase, MGL | |
| 4 | View Details | [502..639] | 208.457575 | Methionine gamma-lyase, MGL |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)