













| General Information: |
|
| Name(s) found: |
ARG2 /
YJL071W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 574 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA |
| Biological Process: |
arginine biosynthetic process
[TAS ornithine biosynthetic process [IMP telomere maintenance [IMP |
| Molecular Function: |
acetyl-CoA:L-glutamate N-acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWRRIFAHEL KYDQPNASSK NLILSVLNTT ATKREAKDYL SKYTNDSGQH NHCLFFIRDL 60
61 HKVAPAILSQ FSSVIKRLGM LGLRPMFVIP PSPTHVNIQA ELLDSIVTEA DLKPLHLKEG 120
121 LTKSRTGLYH SVFSQESRFF DIGNSNFIPI VKPYVYNEET ASEFMTKDVV KFMDCLCQGN 180
181 IPHIDKFFIL NNAGGIPSGE RNDNAHVFIN LSQELEHLSS SLSHNISTLT KREPRSQNLL 240
241 HRMEVYVKKD EISSLECEYH DHLENLLLMD KVLSNLAATA TGLITTVKAA ALSSDRKNPL 300
301 VYNLLTDRSL ISSSLPRFKK KDGEIDSPAN MFDDHAWYEL PSQQVNAAPS NSDAVLVTTV 360
361 LKKGVHIKTY DYKTLTQFNS IGLPKKFHVP EKGAKPSSNS PKLDINKFKS IIDQSFKRSL 420
421 DLHDYIKRIN GKIATIIVIG DYEGIAILTY EGSEENSFVY LDKFAVLPHL KGSLGISDII 480
481 FNLMFKKFPN EILWRSRKDN VVNKWYFQRS VAVLDLSIDL DPEHCDEKQS QFKLFYYGNP 540
541 QYAKRALRDK KRLREFMRSV RDIKPSWENE KNIS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
ARG2 ARG8 CIT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
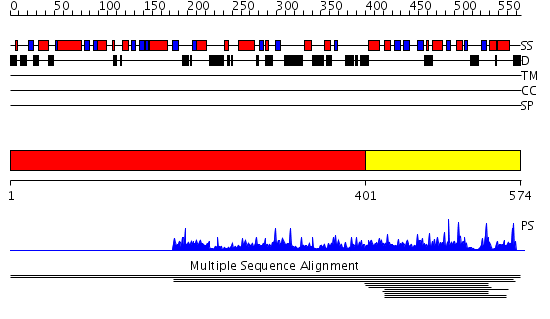
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..400] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [401..574] | 6.009974 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)