













| General Information: |
|
| Name(s) found: |
FAA3 /
YIL009W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 694 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
lipid metabolic process
[IGI N-terminal protein myristoylation [IGI |
| Molecular Function: |
long-chain fatty acid-CoA ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEQHSVAVG KAANEHETAP RRNVRVKKRP LIRPLNSSAS TLYEFALECF NKGGKRDGMA 60
61 WRDVIEIHET KKTIVRKVDG KDKSIEKTWL YYEMSPYKMM TYQELIWVMH DMGRGLAKIG 120
121 IKPNGEHKFH IFASTSHKWM KIFLGCISQG IPVVTAYDTL GESGLIHSMV ETESAAIFTD 180
181 NQLLAKMIVP LQSAKDIKFL IHNEPIDPND RRQNGKLYKA AKDAINKIRE VRPDIKIYSF 240
241 EEVVKIGKKS KDEVKLHPPE PKDLACIMYT SGSISAPKGV VLTHYNIVSG IAGVGHNVFG 300
301 WIGSTDRVLS FLPLAHIFEL VFEFEAFYWN GILGYGSVKT LTNTSTRNCK GDLVEFKPTI 360
361 MIGVAAVWET VRKAILEKIS DLTPVLQKIF WSAYSMKEKS VPCTGFLSRM VFKKVRQATG 420
421 GHLKYIMNGG SAISIDAQKF FSIVLCPMII GYGLTETVAN ACVLEPDHFE YGIVGDLVGS 480
481 VTAKLVDVKD LGYYAKNNQG ELLLKGAPVC SEYYKNPIET AVSFTYDGWF RTGDIVEWTP 540
541 KGQLKIIDRR KNLVKTLNGE YIALEKLESV YRSNSYVKNI CVYADESRVK PVGIVVPNPG 600
601 PLSKFAVKLR IMKKGEDIEN YIHDKALRNA VFKEMIATAK SQGLVGIELL CGIVFFDEEW 660
661 TPENGFVTSA QKLKRREILA AVKSEVERVY KENS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
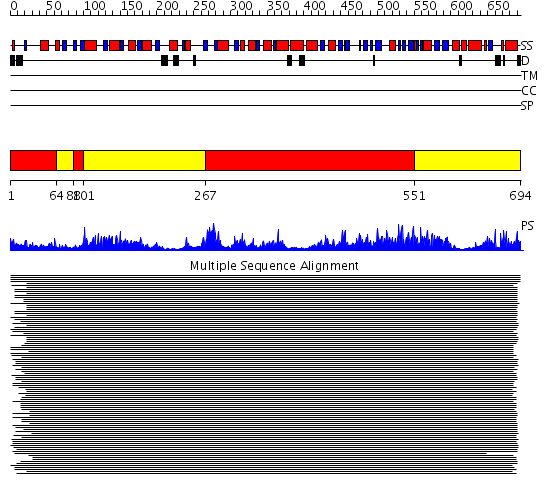
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..63] [88..100] [267..550] |
264.228787 | Luciferase | |
| 2 | View Details | [64..87] [101..266] [551..694] |
264.228787 | Luciferase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)