













| General Information: |
|
| Name(s) found: |
YHR131C
[SGD]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 840 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANNRIERLK SPSSSSTCSM DEVLITSSNN SSSICLETMR QLPREGVSGQ INIIKETAAS 60
61 SSSHAALFIK QDLYEHIDPL PAYPPSYDLV NPNKEVRFPI FGDTAPCPKS SLPPLYAPAV 120
121 YELTLISLKL ERLSPYEISS NRSWRNFIIE INSTQLNFYH IDESLTKHIR NYSSGETKSE 180
181 KEDRIHSDLV HRSDQSQHLH HRLFTLPTRS ASEFKKADQE RISYRVKRDR SRYLTDEALY 240
241 KSFTLQNARF GIPTDYTKKS FVLRMSCESE QFLLRFSHID DMIDWSMYLS IGISVSLDLE 300
301 VREYPDYRIV PRRRRRRRRR RRRRRHTHRS ESSMGSFSQR FIRSNSRPDL IQRYSTGSST 360
361 NNNTTIRERS NTFTAGLLDH YCTGLSKTPT EALISSAASG ESSDNSTLGS TRSLSGCSAS 420
421 RSIASRSLKF KIKNFFRPKN SSRTEKLHRL RSNSSNLNSV IETEEDDEHH ESSGGDHPEP 480
481 GVPVNTTIKV ERPMHRNRAI SMPQRQSLRR AISEEVVPIK FPNSTVGESV HSPSPIEHLS 540
541 VDGCEIMLQS QNAVMKEELR SVASNLVANE RDEASIRPKP QSSSIYLSGL APNGESATDL 600
601 SQSSRSLCLT NRDAEINDDE SATETDEDEN DGETDEYAGD DTNDDTDDSN IGYAYGSESD 660
661 YSCLIEERIR NRRRASSTLS CFSNIPYGTD DIKWKPAIKE ISRRRYLRDS LKCIKPFLDS 720
721 NDCLGKVIYI PVSGPTFETS NKIHFSNRQS LQKQKNHFLK GFIVGPTALI ELNCKNKNAI 780
781 VGTTKDAEDH GEDDGDGDDG EDDDDDDDDD DDDDDDEDDD DDDDDDDDDD DDDDDGQITA 840
841 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
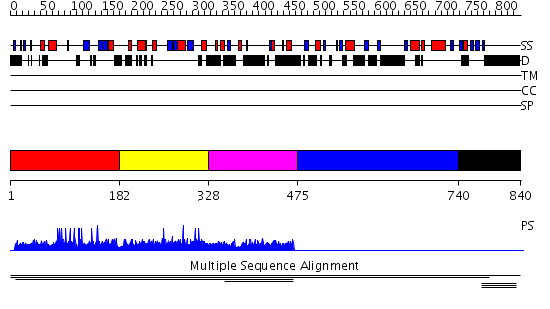
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [182..327] | 4.481486 | PH domain No confident structure predictions are available. | |
| 3 | View Details | [328..474] | 1.003965 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [475..739] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [740..840] | 1.002997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)