













| General Information: |
|
| Name(s) found: |
YHL050C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 697 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
helicase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADTPSVAVQ APPGYGKTEL FHLPLIALAS KGDVEYVSFL FVPYTVLLAN CMIRLGRCGC 60
61 LNVAPVRNFI EEGCDGVTDL YVGIYDDLAS TNFTDRIAAW ENIVECTFRT NNVKLGYLIV 120
121 DELHNFETEV YRQSQFGGIT NLDFDAFEKA IFLSGTAPEA VADAALQRIG LTGLAKKSMD 180
181 INELKRSEDL SRGLSSYPTR MFNLIKEKSK VPLGTNATTT ASTNVRTSAT TTASINVRTS 240
241 ATTTASINVR TSATTTESTN SNTNATTTES TNSSTNATTT ASTNSSTNAT TTESTNASAK 300
301 EDANKDGNAE DNRFHPVTDI NKEPYKRKGS QMVLLERKKL KAQFPNTSEN MNVLQFLGFR 360
361 SDEIKHLFLY GIDIYFCPEG VFTQYGLCKG CQKMFELCVC WAGQKVSYRR MAWEALAVER 420
421 MLRNDEEYKE YLEDIEPYHG DPVGYLKFFS VKRREIYSQI QRNYAWYLAI TRRRETISVL 480
481 DSTRGKQGSQ VFRMSGRQIK ELYYKVWSNL RESKTEVLQY FLNWDEKKCQ EEWEAKDDTV 540
541 FVEALEKVGV FQRLRSMTSA GLQGPQYVKL QFSRHHRQLR SRYELSLGMH LRDQLALGVT 600
601 PSKVPHWTAF LSMLIGLFYN KTFRQKLEYL LEQISEVWLL PHWLDLANVE VLAADNTRVP 660
661 LYMLMVAVHK ELDSDDVPDG RFDIILLCRD SSREVGE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
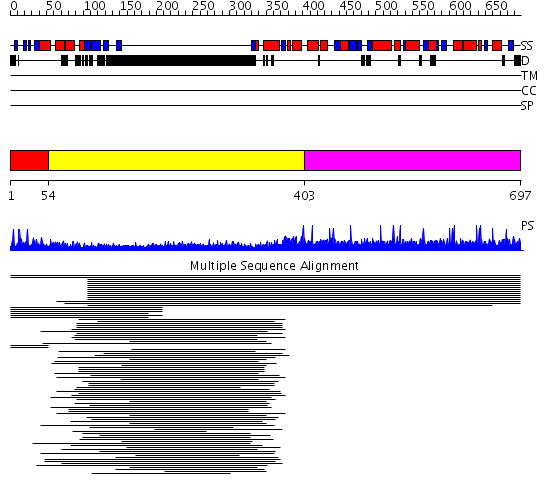
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..53] | 1.030978 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [54..402] | 55.157997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [403..697] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [604..697] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)