













| General Information: |
|
| Name(s) found: |
DRN1 /
YGR093W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 507 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNAKILVAH ISESDADEAI RKIKKVNEKS GPFDLIIIFS NSYDENFELN TDGLPQLILL 60
61 SCDKANNSKS KKINENVTLL HNMGTYKLAN GITLSYFIYP DDTLQGEKKS ILDEFGKSED 120
121 QVDILLTKEW GLSISERCGR LSGSEVVDEL AKKLQARYHF AFSDEINFYE LEPFQWERER 180
181 LSRFLNIPKY GSGKKWAYAF NMPIGDNELK DEPEPPNLIA NPYNSVVTNS NKRPLETETE 240
241 NSFDGDKQVL ANREKNENKK IRTILPSSCH FCFSNPNLED HMIISIGKLV YLTTAKGPLS 300
301 VPKGDMDISG HCLIIPIEHI PKLDPSKNAE LTQSILAYEA SLVKMNYIKF DMCTIVFEIQ 360
361 SERSIHFHKQ VIPVPKYLVL KFCSALDRQV HFNNEKFTRN AKLEFQCYDS HSSKQYVDVI 420
421 NNQSNNYLQF TVYETPEADP KIYLATFNAS ETIDLQFGRR VLAFLLNLPR RVKWNSSTCL 480
481 QTKQQETIEA EKFQKAYRTY DISLTEN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ACO2 DRN1 MRPL16 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
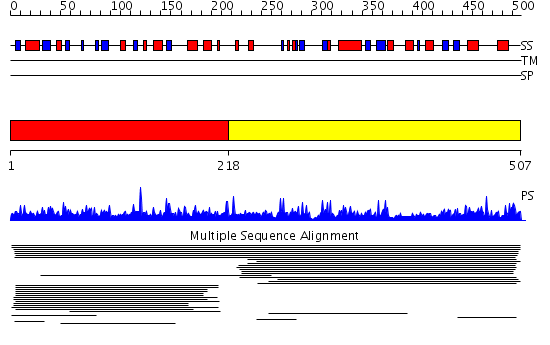
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 9.017997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [218..507] | 9.017939 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [423..507] | 44.113509 | No description for PF04676.5 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)