













| General Information: |
|
| Name(s) found: |
VAB2 /
YEL005C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 282 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVADLTKGIL KWKSRIEFDA VGSSSYYEEL KGLPPLASHK KLTQAAIFNS TKYELLQVKK 60
61 DILSIYEVVS RDIDEERNQM QQIELQLKKS LKKVEHSYKN VLKQRASTNC INGNDRLLAN 120
121 AEKKIGSLNE ELACVNDIVS DIVNNLTALN ANLPKKAQLL KDDSINVAHY PLLFDFLHKS 180
181 CPKSIIATSE ASIHENASPS PLLEHDELPA ESINSFYGEN ELQSDSLAPL QTHDDNISSC 240
241 KKILPPKFNT TSGPSIETNF ENISADGLTY TKCSLKNSIS LT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
VAB2 YDR357C YGL079W YKL061W YLR408C YNL086W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
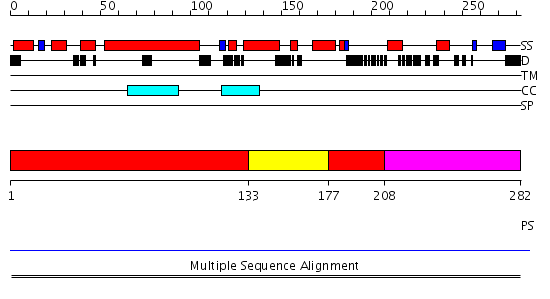
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] [177..207] |
7.03 | Hemolysin E (HlyE, ClyA, SheA) | |
| 2 | View Details | [133..176] | 7.03 | Hemolysin E (HlyE, ClyA, SheA) | |
| 3 | View Details | [208..282] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)