













| General Information: |
|
| Name(s) found: |
SPP41 /
YDR464W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1435 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYDEDDGEI NFNELVGNLL SSHNQEGQEE GEVQGGEQEG DDFEKIYPTS ENIEPKHPDD 60
61 SQHMHNSPDQ NIEIPHFVDE EDELVSVVAN AVQNIDDEQA KPENHLENGS EHVTSDTADD 120
121 NHEKEQQQEW AHILQQEILK SDGEPLRENT ERRVSTSQHH PSQRTDDALD QDDENLRMAI 180
181 LESLQELNTN EEEEKEPEKH EHAAPNDKLS SKKSSKKKKK DKSKNRESSK DKSSKKSKSS 240
241 SHSKKHAKDR NKEKQSKPTN NENTLDLSNI LENLIHENDN AAIDTAKQTV DIQDNSHTDN 300
301 TNNEDVEAQA LVEATLKAFE NELLSSAPTE EPSQEQSIGP VSSRKAVEPP RKPTADDIPL 360
361 AMLQAFKPKK RPPQEKKKTK SKTSKAASTA NKSPASESTS KKKKKKKTVK ESNKSQEAYE 420
421 DDEFSRILAD MVNQVVNTSL KETSTHTATQ DNKLESESDF TSPVQSQYTT EDASTANDDS 480
481 LDLNQIMQNA MAMVFQNQND DEFDENIVED FNRGLGDLSV SDLLPHDNLS RMEKKSVPKS 540
541 SSKSEKKTAI SRRASKKASR DASSVELTEV PSKPKKPSKT EVSLEKKLRK KYVSIANEAA 600
601 SVARKKRWAK NKELKEKEKL ERQTAREERR HKKKLEKQRL AEEQEELKKI VERGPPYPPD 660
661 LRLTKSGKPK KPYRRWTPEE LLKRSQEAEK PRKVKKERKK KEKKMKVPSS ALKKIPLFNF 720
721 VKGNVQPSAR HRLNDIEGSL STIGLHKSPD GVRRILSRPK SEDHEWPLSD SSASQNYDAH 780
781 LKTVVHKEKI PFHPPWTIPS QPPFALPVAR RKKIPNIKKY RKRTNNSFRV SKEGTASTRN 840
841 RILPAILLPI INTLKAAAKS QTAAGATPEE ARKRLATIIQ HAKSTVIRAA LQARKNSMQA 900
901 AHSKGTTTEL ATTASRMKNP LKMIPIFNTS RVKQQLDKQL PARSAGTEIS SSESPDKATP 960
961 DPHSNSTIAG HTLKGVTTPI KIEDSDANVP PVSIAVSTIE PSQDKLELTK RAESVEPVEN1020
1021 NVETAKETQS VQEIKENVGT KASEEVTLTE DKTNGDPKNE KRILIESPVE KTDKKKPGEK1080
1081 IATDLNEDAS LSDKKDGDEK STLHSDAAQL TGNEPDSVNT TTGKPKLIDV SLKPLNEAKP1140
1141 KIPIIFPLKR PQIKPEVSVI NLVQNLVNTK IPEIKNESVD LGSNITDILS STITNILPEI1200
1201 TATDVKNYQY EDENVKYLKK TPRQVLNLDG LVPPSGRCIT KAKRVRRIKK LSADATTAPE1260
1261 ADGKANSESI TYTFDIPSPE EVQSKRSVVL KFAKARLTEA ELSCLKKEIN NVRKRRWREM1320
1321 NSTKNWEYDV KSRLKKRANA FFGEGESETK SKWIEERFQE KVSQEKYKDR LETTETQANN1380
1381 TKIVIDDKEI LNILAVNMNN LNKARCIEKD IQESFREEKL ASLQPKKKRK KSILH |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
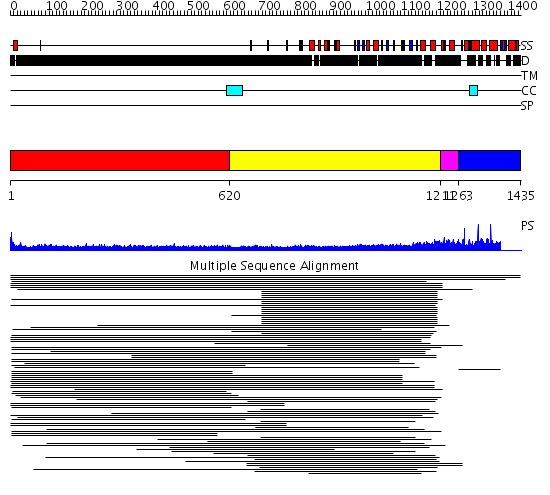
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..619] | 5.162998 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [620..1210] | 20.271997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1211..1262] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [1263..1435] | 1.010962 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [580..664] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [665..875] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [876..983] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [984..1201] | 1.359996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1202..1435] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)