













| General Information: |
|
| Name(s) found: |
RKM4 /
YDR257C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 494 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDFSRDTEN FVCWLKTTAE IEVSPKIEIK DLCCDNQGRA VVATQKIKKD ETLFKIPRSS 60
61 VLSVTTSQLI KDYPSLKDKF LNETGSWEGL IICILYEMEV LQERSRWAPY FKVWNKPSDM 120
121 NALIFWDDNE LQLLKPSLVL ERIGKKEAKE MHERIIKSIK QIGGEFSRVA TSFEFDNFAY 180
181 IASIILSYSF DLEMQDSSVN ENEEEETSEE ELENERYLKS MIPLADMLNA DTSKCNANLT 240
241 YDSNCLKMVA LRDIEKNEQV YNIYGEHPNS ELLRRYGYVE WDGSKYDFGE VLLENIVEAL 300
301 KETFETNTEF LDRCIDILRN NANIQEFLEG EEIVLDSYDC YNNGELLPQL ILLVQILTIL 360
361 CQIPGLCKLD IKAMERQVER IVKKCLQLIE GARATTNCSA TWKRCIMKRL ADYPIKKCVS 420
421 IEKPSKGNSL TREELRDVMA RRVLKSEIDS LQVCEETIDK NYKVIPDEKL LTNILKRKLT 480
481 EEEKSSVKRP CVKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
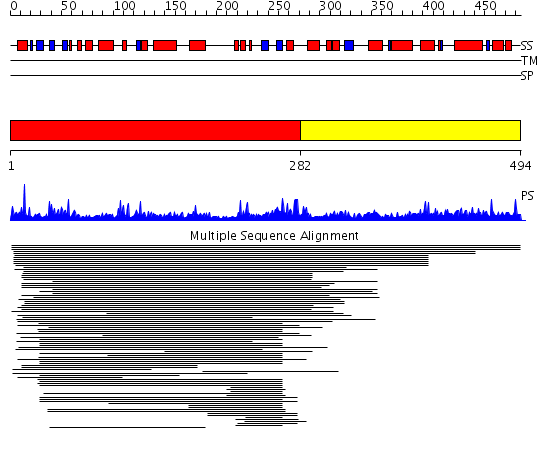
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..281] | 19.062997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [282..494] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)