













| General Information: |
|
| Name(s) found: |
UPC2 /
YDR213W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 913 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI cytoplasm [IDA |
| Biological Process: |
steroid metabolic process
[IMP sterol biosynthetic process [IMP |
| Molecular Function: |
RNA polymerase II transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEVGIQNHK KAVTKPRRRE KVIELIEVDG KKVSTTSTGK RKFHNKSKNG CDNCKRRRVK 60
61 CDEGKPACRK CTNMKLECQY TPIHLRKGRG ATVVKYVTRK ADGSVESDSS VDLPPTIKKE 120
121 QTPFNDIQSA VKASGSSNDS FPSSASTTKS ESEEKSSAPI EDKNNMTPLS MGLQGTINKK 180
181 DMMNNFFSQN GTIGFGSPER LNSGIDGLLL PPLPSGNMGA FQLQQQQQVQ QQSQPQTQAQ 240
241 QASGTPNERY GSFDLAGSPA LQSTGMSLSN SLSGMLLCNR IPSGQNYTQQ QLQYQLHQQL 300
301 QLQQHQQVQL QQYQQLRQEQ HQQVQQQQQE QLQQYQQHFL QQQQQVLLQQ EQQPNDEEGG 360
361 VQEENSKKVK EGPLQSQTSE TTLNSDAATL QADALSQLSK MGLSLKSLST FPTAGIGGVS 420
421 YDFQELLGIK FPINNGNSRA TKASNAEEAL ANMQEHHERA AASVKENDGQ LSDTKSPAPS 480
481 NNAQGGSASI MEPQAADAVS TMAPISMIER NMNRNSNISP STPSAVLNDR QEMQDSISSL 540
541 GNLTKAALEN NEPTISLQTS QTENEDDASR QDMTSKINNE ADRSSVSAGT SNIAKLLDLS 600
601 TKGNLNLIDM KLFHHYCTKV WPTITAAKVS GPEIWRDYIP ELAFDYPFLM HALLAFSATH 660
661 LSRTETGLEQ YVSSHRLDAL RLLREAVLEI SENNTDALVA SALILIMDSL ANASGNGTVG 720
721 NQSLNSMSPS AWIFHVKGAA TILTAVWPLS ERSKFHNIIS VDLSDLGDVI NPDVGTITEL 780
781 VCFDESIADL YPVGLDSPYL ITLAYLDKLH REKNQGDFIL RVFTFPALLD KTFLALLMTG 840
841 DLGAMRIMRS YYKLLRGFAT EVKDKVWFLE GVTQVLPQDV DEYSGGGGMH MMLDFLGGGL 900
901 PSMTTTNFSD FSL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
BUL2 UPC2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
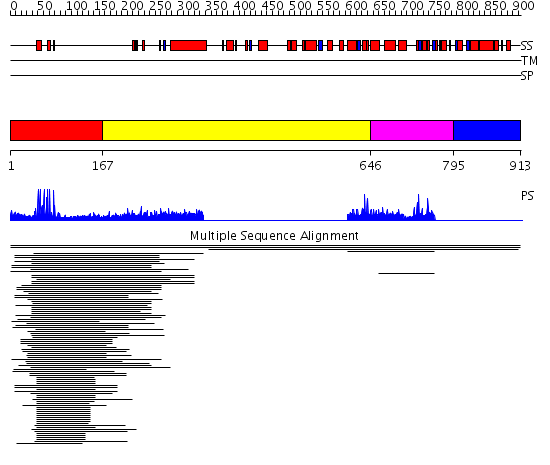
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 46.502279 | PPR1 | |
| 2 | View Details | [167..645] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [646..794] | 1.004965 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [795..913] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [757..913] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.60 |
Source: Reynolds et al. (2008)