













| General Information: |
|
| Name(s) found: |
PGS1 /
YCL004W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 521 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
phospholipid biosynthetic process
[IMP |
| Molecular Function: |
CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTRLLQLTR PHYRLLSLPL QKPFNIKRQM SAANPSPFGN YLNTITKSLQ QNLQTCFHFQ 60
61 AKEIDIIESP SQFYDLLKTK ILNSQNRIFI ASLYLGKSET ELVDCISQAL TKNPKLKVSF 120
121 LLDGLRGTRE LPSACSATLL SSLVAKYGSE RVDCRLYKTP AYHGWKKVLV PKRFNEGLGL 180
181 QHMKIYGFDN EVILSGANLS NDYFTNRQDR YYLFKSRNFS NYYFKLHQLI SSFSYQIIKP 240
241 MVDGSINIIW PDSNPTVEPT KNKRLFLREA SQLLDGFLKS SKQSLPITAV GQFSTLVYPI 300
301 SQFTPLFPKY NDKSTEKRTI LSLLSTITSN AISWTFTAGY FNILPDIKAK LLATPVAEAN 360
361 VITASPFANG FYQSKGVSSN LPGAYLYLSK KFLQDVCRYR QDHAITLREW QRGVVNKPNG 420
421 WSYHAKGIWL SARDKNDANN WKPFITVIGS SNYTRRAYSL DLESNALIIT RDEELRKKMK 480
481 AELDNLLQYT KPVTLEDFQS DPERHVGTGV KIATSILGKK L |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
PGS1 SMK1 YAP7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Widlund PO, et al. (2005) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
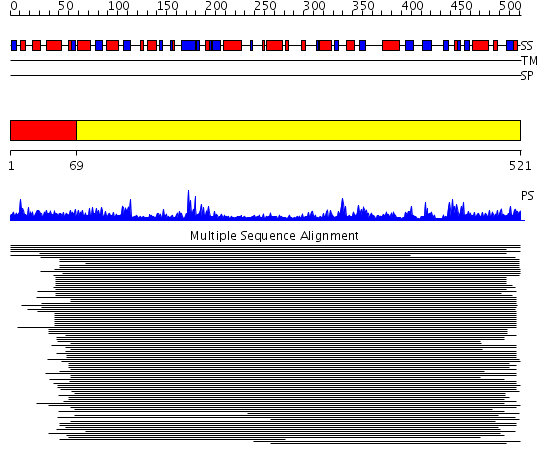
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | 1.113983 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [69..521] | 9.221849 | Phospholipase D |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)