













| General Information: |
|
| Name(s) found: |
HIS7 /
YBR248C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 552 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[TAS |
| Biological Process: |
purine nucleoside monophosphate biosynthetic process
[TAS histidine biosynthetic process [TAS |
| Molecular Function: |
imidazoleglycerol-phosphate synthase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPVVHVIDVE SGNLQSLTNA IEHLGYEVQL VKSPKDFNIS GTSRLILPGV GNYGHFVDNL 60
61 FNRGFEKPIR EYIESGKPIM GICVGLQALF AGSVESPKST GLNYIDFKLS RFDDSEKPVP 120
121 EIGWNSCIPS ENLFFGLDPY KRYYFVHSFA AILNSEKKKN LENDGWKIAK AKYGSEEFIA 180
181 AVNKNNIFAT QFHPEKSGKA GLNVIENFLK QQSPPIPNYS AEEKELLMND YSNYGLTRRI 240
241 IACLDVRTND QGDLVVTKGD QYDVREKSDG KGVRNLGKPV QLAQKYYQQG ADEVTFLNIT 300
301 SFRDCPLKDT PMLEVLKQAA KTVFVPLTVG GGIKDIVDVD GTKIPALEVA SLYFRSGADK 360
361 VSIGTDAVYA AEKYYELGNR GDGTSPIETI SKAYGAQAVV ISVDPKRVYV NSQADTKNKV 420
421 FETEYPGPNG EKYCWYQCTI KGGRESRDLG VWELTRACEA LGAGEILLNC IDKDGSNSGY 480
481 DLELIEHVKD AVKIPVIASS GAGVPEHFEE AFLKTRADAC LGAGMFHRGE FTVNDVKEYL 540
541 LEHGLKVRMD EE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
DUS3 HIS7 YIR035C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
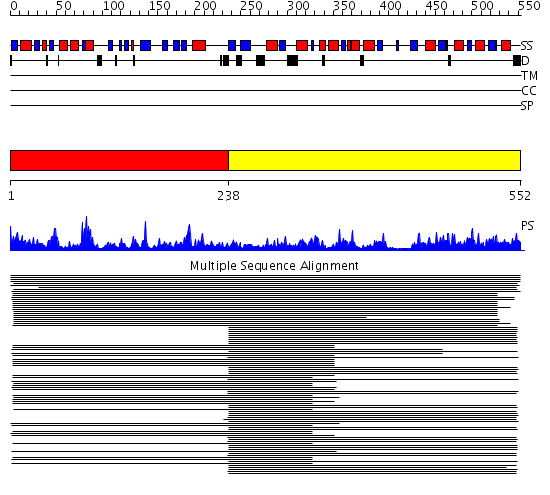
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] | 10161.0 | Cyclase subunit (or domain) of imidazoleglycerolphosphate synthase HisF; GAT subunit, HisH, (or domain) of imidazoleglycerolphosphate synthase HisF | |
| 2 | View Details | [238..552] | 10161.0 | Cyclase subunit (or domain) of imidazoleglycerolphosphate synthase HisF; GAT subunit, HisH, (or domain) of imidazoleglycerolphosphate synthase HisF |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)