













| General Information: |
|
| Name(s) found: |
SWC3 /
YAL011W
[SGD]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 638 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA Swr1 complex [IDA |
| Biological Process: |
chromatin remodeling
[IDA response to drug [IMP histone exchange [IPI endoplasmic reticulum organization [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFGSNARRR QVRMPAVLRT RSKESSIEQK PASRTRTRSR RGKRGRDDDD DDDDEESDDA 60
61 YDEVGNDYDE YASRAKLATN RPFEIVAGLP ASVELPNYNS SLTHPQSIKN SGVLYDSLVS 120
121 SRRTWVQGEM FELYWRRPKK IVSESTPAAT ESPTSGTIPL IRDKMQKMCD CVMSGGPHTF 180
181 KVRLFILKND KIEQKWQDEQ ELKKKEKELK RKNDAEAKRL RMEERKRQQM QKKIAKEQKL 240
241 QLQKENKAKQ KLEQEALKLK RKEEMKKLKE QNKNKQGSPS SSMHDPRMIM NLNLMAQEDP 300
301 KLNTLMETVA KGLANNSQLE EFKKFIEIAK KRSLEENPVN KRPSVTTTRP APPSKAKDVA 360
361 EDHRLNSITL VKSSKTAATE PEPKKADDEN AEKQQSKEAK TTAESTQVDV KKEEEDVKEK 420
421 GVKSEDTQKK EDNQVVPKRK RRKNAIKEDK DMQLTAFQQK YVQGAEIILE YLEFTHSRYY 480
481 LPKKSVVEFL EDTDEIIISW IVIHNSKEIE KFKTKKIKAK LKADQKLNKE DAKPGSDVEK 540
541 EVSFNPLFEA DCPTPLYTPM TMKLSGIHKR FNQIIRNSVS PMEEVVKEME KILQIGTRLS 600
601 GYNLWYQLDG YDDEALSESL RFELNEWEHA MRSRRHKR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
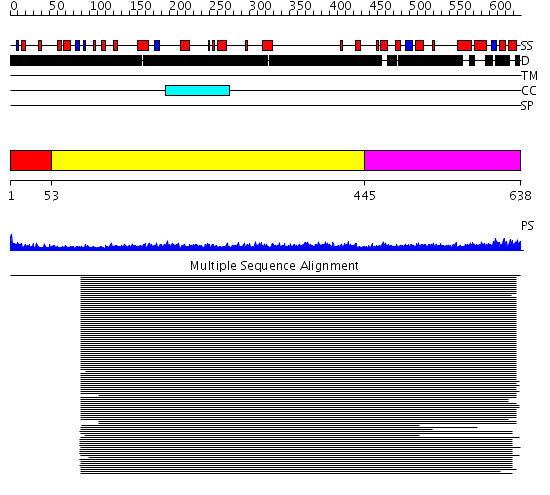
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..52] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [53..444] | 25.69897 | Heavy meromyosin subfragment | |
| 3 | View Details | [445..638] | 8.049995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)