













| General Information: |
|
| Name(s) found: |
AI4 /
Q0065
[SGD]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 544 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[TAS |
| Biological Process: |
RNA splicing
[IDA intron homing [IMP |
| Molecular Function: |
endonuclease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQRWLYSTN AKDIAVLYFM LAIFSGMAGT AMSLIIRLEL AAPGSQYLHG NSQLFNVLVM 60
61 PALIGGFGNY LLPLMIGATD TAFPRINNIA FWVLPMGLVC LVTSTLVESG AGTGWTVYPP 120
121 LSSIQAHSGP SVDLAIFALH LTSISSLLGA INFIVTTLNM RTNGMTMHKL PLFVWSIFIT 180
181 AFLLLLSLPV LSAGITMLLL DRNFNTSFFE VSGGGDPILY EHLFWFFGQT VATIIMLMMY 240
241 NDMHFSKCWK LLKKWITNIM STLFKALFVK MFMSYNNQQD KMMNNTMLKK DNIKRSSETT 300
301 RKMLNNSMNK KFNQWLAGLI DGDGYFGIVS KKYVSLEITV ALEDEMALKE IQNKFGGSIK 360
361 LRSGVKAIRY RLTNKTGMIK LINAVNGNIR NTKRLVQFNK VCILLGIDFI YPIKLTKDNS 420
421 WFVGFFDADG TINYSFKNNH PQLTISVTNK YLQDVQEYKN ILGGNIYFDK SQNGYYKWSI 480
481 QSKDMVLNFI NDYIKMNPSR TTKMNKLYLS KEFYNLKELK AYNKSSDSMQ YKAWLNFENK 540
541 WKNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
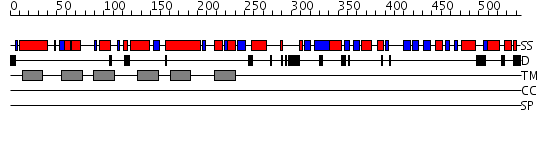
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..425] | 135.0 | Mitochondrial cytochrome c oxidase, subunit I | |
| 2 | View Details | [426..544] | 9.221849 | The structure and DNA recognition of a bifunctional homing endonuclease and group I intron splicing factor |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|