













| General Information: |
|
| Name(s) found: |
h0004690
[Lars HPF FASTA]
|
| Description(s) found: |
|
| Organism: | SPECIES UNKNOWN |
| Length: | 1303 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPGPRGAAGG LAPEMRGAGA AGLLALLLLL LLLLLGLGGR VEGGPAGERG AGGGGALARE 60
61 RFKVVFAPVI CKRTCLKGQC RDSCQQGSNM TLIGENGHST DTLTGSGFRV VVCPLPCMNG 120
121 GQCSSRNQCL CPPDFTGRFC QVPAGGAGGG TGGSGPGLSR TGALSTGALP PLAPEGDSVA 180
181 SKHAIYAVQV IADPPGPGEG PPAQHAAFLV PLGPGQISAE VQAPPPVVNV RVHHPPEASV 240
241 QVHRIESSNA ESAAPSQHLL PHPKPSHPRP PTQKPLGRCF QDTLPKQPCG SNPLPGLTKQ 300
301 EDCCGSIGTA WGQSKCHKCP QLQYTGVQKP GPVRGEVGAD CPQGYKRLNS THCQDINECA 360
361 MPGVCRHGDC LNNPGSYRCV CPPGHSLGPS RTQCIADKPE EKSLCFRLVS PEHQCQHPLT 420
421 TRLTRQLCCC SVGKAWGARC QRCPTDGTAA FKEICPAGKG YHILTSHQTL TIQGESDFSL 480
481 FLHPDGPPKP QQLPESPSQA PPPEDTEEER GVTTDSPVSE ERSVQQSHPT ATTTPARPYP 540
541 ELISRPSPPT MRWFLPDLPP SRSAVEIAPT QVTETDECRL NQNICGHGEC VPGPPDYSCH 600
601 CNPGYRSHPQ HRYCVDVNEC EAEPCGPGRG ICMNTGGSYN CHCNRGYRLH VGAGGRSCVD 660
661 LNECAKPHLC GDGGFCINFP GHYKCNCYPG YRLKASRPPV CEDIDECRDP SSCPDGKCEN 720
721 KPGSFKCIAC QPGYRSQGGG ACRDVNECAE GSPCSPGWCE NLPGSFRCTC AQGYAPAPDG 780
781 RSCLDVDECE AGDVCDNGIC SNTPGSFQCQ CLSGYHLSRD RSHCEDIDEC DFPAACIGGD 840
841 CINTNGSYRC LCPQGHRLVG GRKCQDIDEC SQDPSLCLPH GACKNLQGSY VCVCDEGFTP 900
901 TQDQHGCEEV EQPHHKKECY LNFDDTVFCD SVLATNVTQQ ECCCSLGAGW GDHCEIYPCP 960
961 VYSSAEFHSL CPDGKGYTQD NNIVNYGIPA HRDIDECMLF GSEICKEGKC VNTQPGYECY1020
1021 CKQGFYYDGN LLECVDVDEC LDESNCRNGV CENTRGGYRC ACTPPAEYSP AQRQCLSPEE1080
1081 MDVDECQDPA ACRPGRCVNL PGSYRCECRP PWVPGPSGRD CQLPESPAER APERRDVCWS1140
1141 QRGEDGMCAG PLAGPALTFD DCCCRQGRGW GAQCRPCPPR GAGSHCPTSQ SESNSFWDTS1200
1201 PLLLGKPPRD EDSSEEDSDE CRCVSGRCVP RPGGAVCECP GGFQLDASRA RCVDIDECRE1260
1261 LNQRGLLCKS ERCVNTSGSF RCVCKAGFAR SRPHGACVPQ RRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
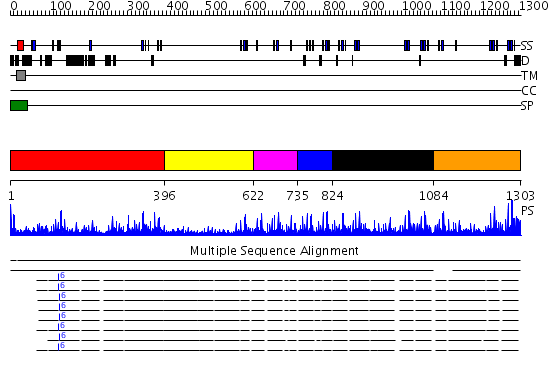
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..395] | 15.69897 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor | |
| 2 | View Details | [396..621] | 3.08 | Crystal structure of Methylated Pvs25, an ookinete protein from Plasmodium vivax | |
| 3 | View Details | [622..734] | 5.0 | Fibrillin-1 | |
| 4 | View Details | [735..823] | 3.69897 | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, MINIMIZED AVERAGE STRUCTURE | |
| 5 | View Details | [824..1083] | 4.29 | Crystal structure of Methylated Pvs25, an ookinete protein from Plasmodium vivax | |
| 6 | View Details | [1084..1303] | 5.522879 | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, holo form. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|