













| General Information: |
|
| Name(s) found: |
TEB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 2154 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
photomorphogenesis
[IMP]
DNA replication [IGI] DNA recombination [IGI] regulation of gene expression [IGI] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] DNA binding [IEA] DNA-directed DNA polymerase activity [IEA] ATP-dependent helicase activity [IEA] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSDSSKSRI DQFYVSKKRK HQSPNLKSGR NEKNVKVTGE RSPGDKGTLD SYLKASLDDK 60
61 STTNSGLQAR QEAFTRKLDL EVSASSVGQN IHPCLPKPVS FATFKECLGQ NGSQDLHKEG 120
121 VAAETHATDG LLCANQKDNS ELRDFATSFL SLYCSGVQSV VGSPPHQKEN ELKRRSSSSS 180
181 LAQDIQISHK RRCESENIPS LDDLTNPLGS KPESLARNGN NRDKPVSDPT KKMPSNESVE 240
241 IPMGLRKCSK APESSAHLTE FHTPGSAIKS CPVGTPKSGC GSSMFSPGEA FWNEAIQVAD 300
301 GLTIPIENFG SVEAKVRDQH VTILSCSKKT DKCTEKLERS LDLDEIRVKD KDAIGFSKVV 360
361 EKHGRDFNKE VYQLPVKNLE LLFQDKNING GIQERCASFD QNNITLGSSR ISESAFVGNK 420
421 GCENLDIANN AQADKGLIGK MYPEPEGKKV LLCEENRGVR SVSMISNMRK PVGSSESEES 480
481 HTPSSSHRNY DGLSLSTWLP SEVCSVYNKK GISKLYPWQV ECLQVDGVLQ KRNLVYCAST 540
541 SAGKSFVAEV LMLRRVIRTG KMALLVLPYV SICAEKAEHL EVLLEPLGKH VRSYYGNQGG 600
601 GTLPKDTSVA VCTIEKANSL INRLLEEGRL SELGIIVIDE LHMVGDQHRG YLLELMLTKL 660
661 RYAAGEGSSE SSSGESSGTS SGKADPAHGL QIVGMSATMP NVGAVADWLQ AALYQTEFRP 720
721 VPLEEYIKVG STIYNKKMEV VRTIPKAADM GGKDPDHIVE LCNEVVQEGN SVLIFCSSRK 780
781 GCESTARHIS KLIKNVPVNV DGENSEFMDI RSAIDALRRS PSGVDPVLEE TLPSGVAYHH 840
841 AGLTVEEREI VETCYRKGLV RVLTATSTLA AGVNLPARRV IFRQPMIGRD FIDGTRYKQM 900
901 SGRAGRTGID TKGDSVLICK PGELKRIMAL LNETCPPLQS CLSEDKNGMT HAILEVVAGG 960
961 IVQTAKDIHR YVRCTLLNST KPFQDVVKSA QDSLRWLCHR KFLEWNEETK LYTTTPLGRG1020
1021 SFGSSLCPEE SLIVLDDLLR AREGLVMASD LHLVYLVTPI NVGVEPNWEL YYERFMELSP1080
1081 LEQSVGNRVG VVEPFLMRMA HGATVRTLNR PQDVKKNLRG EYDSRHGSTS MKMLSDEQML1140
1141 RVCKRFFVAL ILSKLVQEAS VTEVCEAFKV ARGMVQALQE NAGRFSSMVS VFCERLGWHD1200
1201 LEGLVAKFQN RVSFGVRAEI VELTSIPYIK GSRARALYKA GLRTSQAIAE ASIPEIVKAL1260
1261 FESSAWAAEG TGQRRIHLGL AKKIKNGARK IVLEKAEEAR AAAFSAFKSL GLDVNELSKP1320
1321 LPLAPASSLN GQETTERDIS RGSVGPDGLQ QSIEGHMECE NFDMDNHREK PSEVLGDATL1380
1381 GVSSEINLTS RLPNFRPIGT AVGTNGPSAV SILSSDTFPI PVYDNREIKP KDNVEQHLTR1440
1441 NDHIPLSSNK DGTGEKGPVT AGNISGGFDS FLELWGSAGE FFFDLHYNKL QDLNSRISYE1500
1501 IHGIAICWNC SPVYYVNLNK DLPNLECVEK QKLIEDAVIG KSEVLASHNM LDVIKSRWNK1560
1561 ISKIMGNVNT RKFTWNLKVQ IQVLKSPAIS IQRCTRLNLP EGIRDELVDG SWLMMPPLHT1620
1621 SHTIDMSIVI WILWPDEERH SNPNIDKEVK KRLSPEAAEA ANRSGRWRNQ IRRVAHNGCC1680
1681 RRVAQTRALC SALWKILVSE ELLQALTTIE MPLVNVLADM ELWGIGIDIE GCLRARNILR1740
1741 DKLRSLEKKA FELAGMTFSL HNPADIANVL FGQLKLPIPE NQSKGKLHPS TDKHCLDLLR1800
1801 NEHPVVPIIK EHRTLAKLLN CTLGSICSLA KLRLSTQRYT LHGRWLQTST ATGRLSIEEP1860
1861 NLQSVEHEVE FKLDKNGRDV SSDADRYKIN ARDFFVPTQE NWLLLTADYS QIELRLMAHF1920
1921 SRDSSLISKL SQPEGDVFTM IAAKWTGKAE DSVSPHDRDQ TKRLIYGILY GMGANRLAEQ1980
1981 LECTSDEAKE KIRSFKSSFP AVTSWLNETI SFCQEKGYIQ TLKGRRRFLS KIKFGNAKEK2040
2041 SKAQRQAVNS MCQGSAADII KIAMINIYSA IAEDVDTAAS SSSSETRFHM LKGRCRILLQ2100
2101 VHDELVLEVD PSYVKLAAML LQTSMENAVS LLVPLHVKLK VGKTWGSLEP FQTD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
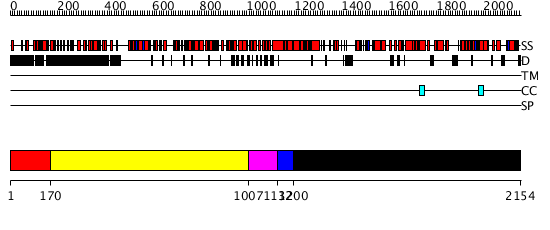
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | 1.264996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [170..1006] | 29.0 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [1007..1131] | 70.39794 | No description for 2p6rA was found. | |
| 4 | View Details | [1132..1199] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1200..2154] | 1000.0 | 5' to 3' exonuclease domain of DNA polymerase Taq; Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)