













| General Information: |
|
| Name(s) found: |
PUB1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSAQSRRI RVTIVAADGL YKRDVFRFPD PFAVLTVDGE QTHTTTAIKK TLNPYWNETF 60
61 EVNVTDNSTI AIQVFDQKKF KKKGQGFLGV INLRVGDVLD LAIGGDEMLT RDLKKSNENT 120
121 VVHGKIIINL STTAQSTLQV PSSAASGART QRTSITNDPQ SSQSSSVSRN PASSRAGSPT 180
181 RDNAPAASPA SSEPRTFSSF EDQYGRLPPG WERRTDNLGR TYYVDHNTRS TTWIRPNLSS 240
241 VAGAAAAELH SSASSANVTE GVQPSSSNAA RRTEASVLTS NATTAGSGEL PPGWEQRYTP 300
301 EGRPYFVDHN TRTTTWVDPR RQQYIRSYGG PNNATIQQQP VSQLGPLPSG WEMRLTNTAR 360
361 VYFVDHNTKT TTWDDPRLPS SLDQNVPQYK RDFRRKLIYF LSQPALHPLP GQCHIKVRRN 420
421 HIFEDSYAEI MRQSATDLKK RLMIKFDGED GLDYGGLSRE YFFLLSHEMF NPFYCLFEYS 480
481 SVDNYTLQIN PHSGINPEHL NYFKFIGRVI GLAIFHRRFV DAFFVVSFYK MILQKKVTLQ 540
541 DMESMDAEYY RSLVWILDND ITGVLDLTFS VEDNCFGEVV TIDLKPNGRN IEVTEENKRE 600
601 YVDLVTVWRI QKRIEEQFNA FHEGFSELIP QELINVFDER ELELLIGGIS EIDMEDWKKH 660
661 TDYRSYSEND QIIKWFWELM DEWSNEKKSR LLQFTTGTSR IPVNGFKDLQ GSDGPRKFTI 720
721 EKAGEPNKLP KAHTCFNRLD LPPYTSKKDL DHKLSIAVEE TIGFGQE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
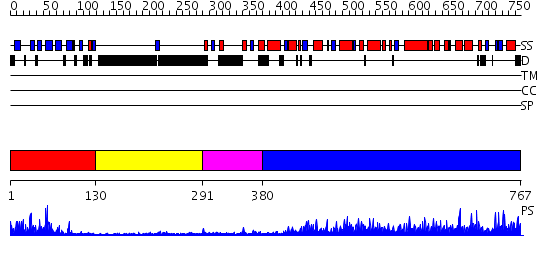
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 19.39794 | No description for 2jqzA was found. | |
| 2 | View Details | [130..290] | 2.48 | NMR structure of WW domains (WW3-4) from Suppressor of Deltex | |
| 3 | View Details | [291..379] | 5.0 | NMR structure of WW domains (WW3-4) from Suppressor of Deltex | |
| 4 | View Details | [380..767] | 140.0 | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)