













| General Information: |
|
| Name(s) found: |
nhr-23 /
CE39086
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
molting cycle, protein-based cuticle
[IMP locomotory behavior [IMP nematode larval development [IMP post-embryonic body morphogenesis [IMP regulation of transcription, DNA-dependent [IEA oviposition [IMP positive regulation of multicellular organism growth [IMP growth [IMP reproduction [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQAVEQKLAD ATRTLHAKSS LQPSLSIETP KSKENDESGC ESSNCMFHPH TIKSEPNFCF 60
61 AREFKSVPDD FRIGGGDLQM GNNSKRLTCV IDTNRVDMAG ILPDNMSFRG LPENKSLLVS 120
121 AQIEVIPCKV CGDKSSGVHY GVITCEGCKG FFRRSQSSIV NYQCPRQKNC VVDRVNRNRC 180
181 QYCRLKKCIE LGMSRDAVKF GRMSKKQREK VEDEVQVRMH KELAANGLGY QAIYGDYSPP 240
241 PSHPSYCFDQ SMYGHYPSGT STPVNGYSIA VAATPTTPMP QNMYGATPSS TNGTQYVAHQ 300
301 ATGGSFPSPQ VPEEDVATRV IRAFNQQHSS YTTQHGVCNV DPDCIPHLSR AGGWELFARE 360
361 LNPLIQAIIE FAKSIDGFMN LPQETQIQLL KGSVFELSLV FAAMYYNVDA QAVCGERYSV 420
421 PFACLIAEDD AEMQLIVEVN NTLQEIVHLQ PHQSELALLA AGLILEQVSS SHGIGILDTA 480
481 TIATAETLKN ALYQSVMPRI GCMEDTIHRI QDVETRIRQT ARLHQEALQN FRMSDPTSSE 540
541 KLPALYKELF TADRP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
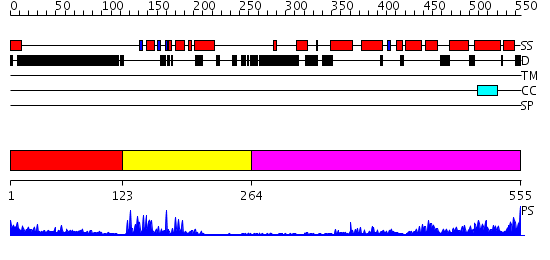
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 14.457997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [123..263] | 25.09691 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 3 | View Details | [264..555] | 54.154902 | LIGAND-BINDING DOMAIN OF THE HUMAN NUCLEAR RECEPTOR RXR-ALPHA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)