













| General Information: |
|
| Name(s) found: |
pig-1 /
CE40473
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 703 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
asymmetric cell division
[IMP generation of neurons [IMP cell fate specification [IMP protein amino acid phosphorylation [IEA |
| Molecular Function: |
magnesium ion binding
[IEA ATP binding [IEA protein tyrosine kinase activity [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKYEVLQGF YAVHDELGSG GFGKVRLATH LLTNQKVAIK IIDKKQLGHD LPRVQTEMDA 60
61 LRNLSHQNIC RLYHYIETED KFFIVMEYCS GGEMFDYIVR KERLEESEAR HFFRQLVSAI 120
121 AFVHSQGYAH RDLKPENLLL TEDLHLKLID FGLCAKTEKG RIDKHNLDTC CGSPAYAAPE 180
181 LIQGLQYKGN EADVWSMGIL LYTLLVGALP FEDDNMQIMY KKIQSGCFYE PEFLSPLSKQ 240
241 LLRAMLQVVP ERRISVKKLL EHDWLNHKYT QPVKWNTIYD KNFIDRDVAR VMSKYYGFES 300
301 TDKMIEKIKE WNFDYMTSTY YALLHRKRNG MEIILPMVRN STNTAPPNVQ NILCSPTIHA 360
361 SLENNLDKSG LEDDDSDPSS ISSSSDISAR LKKNCVVSDE SSSSRFVKPM SPAAEKDKKM 420
421 SYVNAMLTMP SQFTGRSPLR IPESPMSVRS SDSASLGSAA TPSRGGVKDN DKENASTGKN 480
481 YRMGASTCKS RGPLKITGVE EGTMKSVYTT PNTRPTLRGL FSPGNAEHKK RQRARSSDRA 540
541 SIGMPPGSPV SIGSAHSANN ADGRTPRSRI KTNRLPQRVF TSLERKKEKL ITLLTPRKMQ 600
601 RDSPQVLKDV KNMVNVSMTA SQDPEEVRNL LKKVFDDERM RYELNGWKFL ATQETVHGWM 660
661 TVELEIVRLQ MFDKVGIRRK RLKGDAFMYK KVCEKILQMA KIE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
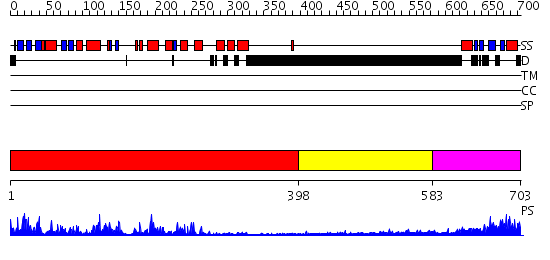
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..397] | 85.0 | No description for 2qnjA was found. | |
| 2 | View Details | [398..582] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [583..703] | 35.30103 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)