













| General Information: |
|
| Name(s) found: |
VHAA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 817 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuole
[IDA]
trans-Golgi network transport vesicle membrane [IDA] |
| Biological Process: |
ATP synthesis coupled proton transport
[IEA]
|
| Molecular Function: |
ATPase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEFLDKLPQ MDLMRSEKMT LVQLIIPVES AHRSITYLGE LGLLQFRDLN ADKSPFQRTF 60
61 ANQVKRCGEM SRKLRFFKDQ IDKAGLRCSP RLEIEPDIAL GDLERQLADH EHEVLEMNSN 120
121 SEKLRQTYNE LLEFKIVLEK ASGFLVSSNT HAIGEEIELH ESTYSNNGFI ETASLLEQEM 180
181 NPGHSNQSGL RFISGIINKD KLLKFERMLF RATRGNMLFN QTTSDEEIMD PSTSEMVEKV 240
241 VFVVFFSGEQ ARTKILKICE AFGANCYPVP EDTTKQRQLT REVLSRLSDL EATLDAGTRH 300
301 RNNALNSVGY SLTNWITTVR REKAVYDTLN MLNFDVTKKC LVGEGWCPTF AKTQIHEVLQ 360
361 RATFDSSSQV GVIFHVMQAV ESPPTYFRTN KLTNAFQEII DAYGVARYQE ANPAVYSVVT 420
421 YPFLFAVMFG DWGHGLCLLL GALYLLARER KLSTQKLGSF MEMLFGGRYV ILLMALFSIY 480
481 CGLIYNEFFS VPFHIFGGSA YKCRDTTCSD AYTVGLIKYR DPYPFGVDPS WRGSRTELPY 540
541 LNSLKMKMSI LLGIAQMNLG LILSFFNARF FGSSLDIRYQ FIPQMIFLNS LFGYLSLLII 600
601 IKWCTGSQAD LYHVMIYMFL SPTEELGENE LFWGQRPLQI VLLLLAFIAV PWMLFPKPFA 660
661 LRKIHMERFQ GRTYGVLVSS EVDLDVEPDS ARGGGHHEEE FNFSEIFVHQ LIHSIEFVLG 720
721 SVSNTASYLR LWALSLAHSE LSTVFYEKVL LLAWGYENIL IRLIGVAVFA FATAFILLMM 780
781 ETLSAFLHAL RLHWVEFMGK FFNGDGYKFK PFSFALI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
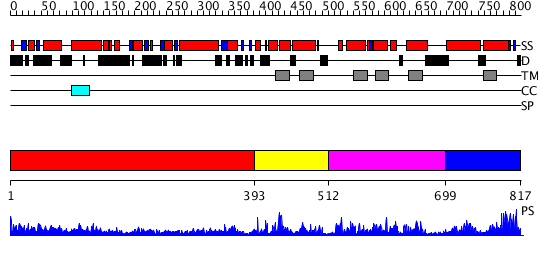
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..392] | 23.045757 | Heavy meromyosin subfragment | |
| 2 | View Details | [393..511] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [512..698] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [699..817] | 1.125972 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|