













| General Information: |
|
| Name(s) found: |
NMT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 434 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
ribosome [IDA] |
| Biological Process: |
embryonic shoot morphogenesis
[IMP]
N-terminal protein myristoylation [IDA] growth [IMP] |
| Molecular Function: |
glycylpeptide N-tetradecanoyltransferase activity
[IDA]
myristoyltransferase activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADNNSPPGS VEQKADQIVE ANPLVKDDTS LETIVRRFQD SMSEAKTHKF WETQPVGQFK 60
61 DIGDTSLPEG PIEPATPLSE VKQEPYNLPS VYEWTTCDMN SDDMCSEVYN LLKNNYVEDD 120
121 ENMFRFNYSK EFLRWALRPP GYYQSWHIGV RAKTSKKLVA FISGVPARIR VRDEVVKMAE 180
181 INFLCVHKKL RSKRLAPVMI KEVTRRVHLE NIWQAAYTAG VILPTPITTC QYWHRSLNPK 240
241 KLIDVGFSRL GARMTMSRTI KLYKLPDAPI TPGFRKMEPR DVPAVTRLLR NYLSQFGVAT 300
301 DFDENDVEHW LLPREDVVDS YLVESPETHD VTDFCSFYTL PSTILGNPNY TTLKAAYSYY 360
361 NVATQTSFLQ LMNDALIVSK QKGFDVFNAL DVMHNESFLK ELKFGPGDGQ LHYYLYNYRL 420
421 KSALKPAELG LVLL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
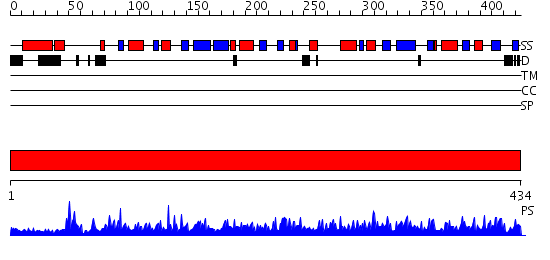
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..434] | 1000.0 | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)