













| General Information: |
|
| Name(s) found: |
gi|50944003
[NCBI NR]
gi|35215184 [NCBI NR] gi|35215045 [NCBI NR] gi|215694402 [NCBI NR] gi|115475838 [NCBI NR] gi|113623484 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 660 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAQVQAVPA AEGGGAPPQA NGVVAAGSAA AAAATFQATS LYVGDLDVSV QDAQLFDVFS 60
61 QVGSVVSVRV CRDVNTRLSL GYAYVNFSSP ADAARALEML NFTPINGKPI RIMYSNRDPS 120
121 SRKSGAANIF IKNLDKSIDN KALYDTFSVF GNILSCKVAT EMSGESKGYG FVQFELEEAA 180
181 QNAISKLNGM LLNDKKVYVG PFVRKQEREN VSGNPKFNNV YVKNLSESTT EDNLKEIFGK 240
241 FGPITSVVVM REGDGKSRCF GFVNFENPDD AARAVEDLNG KKFDDKEWYV CRAQKKSERE 300
301 MELKEKFEKN IKEAADKNQG TNLYLKNLDD SIDDDEKLKE IFADFGTITS CKVMRDLNGV 360
361 SKGSGFVAFK SAEDASRALV AMNGKMIGSK PLYVALAQRK EERRARLQAQ FSQMRPMVMP 420
421 PSVAPRMPMY PPGVPGVGQQ LFYGQPPPAF VNPQPGFGFQ QHLIPGMRPS VGPIPNFVMP 480
481 MVQQGQQPQR PAGRRAGTGG IQQPMPMGHQ QMLPRGSRGG YRYASGRGMP DNAFRGVGGL 540
541 VPSPYEMGRM PLSDAGAPPQ VPIGALASAL ANSPPDQQRL MLGESLYPLV DQLEHDQAAK 600
601 VTGMLLEMDQ TEVLHLIESP DALKAKVAEA MEVLRNAQQQ QANTPTDQLA ALTLSDGVVS 660
661 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
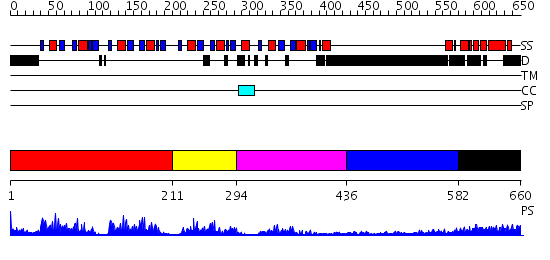
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 56.09691 | Poly(A)-binding protein | |
| 2 | View Details | [211..293] | 3.39794 | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | |
| 3 | View Details | [294..435] | 4.74 | No description for 2f3jA was found. | |
| 4 | View Details | [436..581] | 5.522879 | Sec24 | |
| 5 | View Details | [582..660] | 17.0 | No description for 2dydA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)