













| General Information: |
|
| Name(s) found: |
ROC1_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 784 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPARRMPPV IGRNGVAYES PSAQLPLTQA DMLDSHHLQQ ALQQQYFDQI PVTTTAAADS 60
61 GDNMLHGRAD AGGLVDEFES KSCSENVDGA GDGLSGDDQD PNQRPRKKRY HRHTQHQIQE 120
121 MEAFFKECPH PDDKQRKELS RELGLEPLQV KFWFQNKRTQ MKNQHERHEN AQLRAENDKL 180
181 RAENMRYKEA LSSASCPNCG GPAALGEMSF DEHHLRVENA RLRDEIDRIS GIAAKHVGKP 240
241 PIVSFPVLSS PLAVAAARSP LDLAGAYGVV TPGLDMFGGA GDLLRGVHPL DADKPMIVEL 300
301 AVAAMDELVQ MAQLDEPLWS SSSEPAAALL DEEEYARMFP RGLGPKQYGL KSEASRHGAV 360
361 VIMTHSNLVE ILMDVNQFAT VFSSIVSRAS THEVLSTGVA GNYNGALQVM SMEFQVPSPL 420
421 VPTRESYFVR YCKNNSDGTW AVVDVSLDSL RPSPVQKCRR RPSGCLIQEM PNGYSKVTWV 480
481 EHVEVDDSSV HNIYKPLVNS GLAFGAKRWV GTLDRQCERL ASAMASNIPN GDLGVITSVE 540
541 GRKSMLKLAE RMVASFCGGV TASVAHQWTT LSGSGAEDVR VMTRKSVDDP GRPPGIVLNA 600
601 ATSFWLPVPP AAVFDFLRDE TSRSEWDILS NGGAVQEMAH IANGRDHGNS VSLLRVNSAN 660
661 SNQSNMLILQ ESCTDASGSY VVYAPVDIVA MNVVLNGGDP DYVALLPSGF AILPDGPSGN 720
721 AQAAVGENGS GSGGGSLLTV AFQILVDSVP TAKLSLGSVA TVNSLIACTV ERIKAAVCRD 780
781 SNPQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
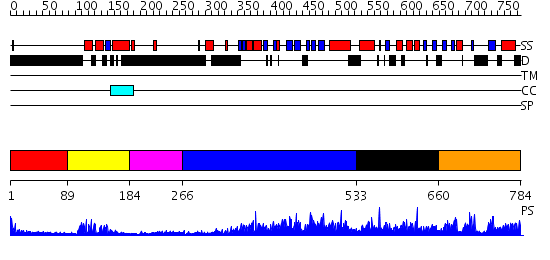
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..183] | 17.221849 | Paired protein | |
| 3 | View Details | [184..265] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [266..532] | 2.73 | No description for 2r55A was found. | |
| 5 | View Details | [533..659] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [660..784] | 2.064973 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)