













| General Information: |
|
| Name(s) found: |
gi|4995952
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 881 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLLSSIEQA CDICRLKKLK CSKEKPKCAK CLKNNWECRY SPKTKRSPLT RAHLTEVESR 60
61 LERLEQLFLL IFPREDLDMI LKMDSLQDIK ALLTGLFVQD NVNKDAVTDR LASVETDMPL 120
121 TLRQHRISAT SSSEESSNKG QRQLTVSIDS AAHHDNSTIP LDFMPRDALH GFDWSEEDDM 180
181 SDGLPFLKTD PNNNGFFGDG SLLCILRSIG FKPENYTNSN VNRLPTMITD RYTLASRSTT 240
241 SRLLQSYLNN FHPYCPIVHS PTLMMLYNNQ IEIASKDQWQ ILFNCILAIG AWCIEGESTD 300
301 IDVFYYQNAK SHLTSKVFES GSIILVTALH LLSRYTQWRQ KTNTSYNFHS FSIRMAISLG 360
361 LNRDLPSSFS DSSILEQRRR IWWSVYSWEI QLSLLYGRSI QLSQNTISFP SSVDDVQRTT 420
421 TGPTIYHGII ETARLLQVFT KIYELDKTVT AEKSPICAKK CLMICNEIEE VSRQAPKFLQ 480
481 MDISTTALTN LLKEHPWLSF TRFELKWKQL SLIIYVLRDF FTNFTQKKSQ LEQDQNDHQS 540
541 YEVKRCSIML SDAAQRTVMS VSSYMDNHNV TPYFAWNCSY YLFNAVLVPI KTLLSNSKSN 600
601 AENNETAQLL QQINTVLMLL KKLATFKIQT CEKYIQVLEE VCAPFLLSQC AIPLPHISYN 660
661 NSNGSAIKNI VGSATIAQYP TLPEENVNNI SVKYVSPGSV GPSPVPLKSG ASFSDLVKLL 720
721 SNRPPSRNSP VTIPRSTPSH RSVTPFLGQQ QQLQSLVPLT PSALFGGANF NQSGNIADSS 780
781 LSFTFTNSSN GPNLITTQTN SQALSQPIAS SNVHDNFMNN EITASKIDDG NNSKPLSPGW 840
841 TDQTAYNAFG ITTGMFNTTT MDDVYNYLFD DEDTPPNPKK E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
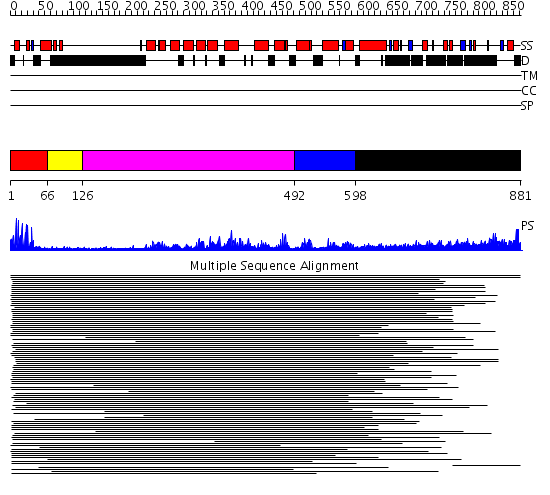
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | 335.31133 | Gal4; CD2-Gal4 | |
| 2 | View Details | [66..125] | 18.958607 | Gal4-like dimerisation domain No confident structure predictions are available. | |
| 3 | View Details | [126..491] | 4.211986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [492..597] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [598..881] | 11.100997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [655..778] | 2.736996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [779..881] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)