













| General Information: |
|
| Name(s) found: |
PNP_CAUCR
[Swiss-Prot]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Caulobacter vibrioides |
| Length: | 712 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFDIKRKTIE WGGKTLVLET GRIARQADGA VLATMGETVV LATAVFAKSQ KPGQDFFPLT 60
61 VNYQEKTFAA GKIPGGFFKR EGRPSEKETL VSRLIDRPIR PLFVKGFKNE VQVVVTVLQH 120
121 DLENDPDILG MVAASAALCL SGAPFMGPIG AARVGWVDGA YVLNPTLDEM KESKMDLVVA 180
181 GTADAVMMVE SEIQELSEEI VLGGVNFAHQ QMQAVIDAII DLAEHAAKEP FAFEPEDTDA 240
241 IKAKMKDLVG ADIAAAYKIQ KKQDRYEAVG AAKKKAIAAL GLSDENPTGY DPLKLGAIFK 300
301 ELEADVVRRG ILDTGLRIDG RDVKTVRPIL GEVGILPRTH GSALFTRGET QAIVVATLGT 360
361 GDDEQFIDAL EGTYKESFLL HYNFPPYSVG ETGRMGSPGR REIGHGKLAW RALRPMLPTK 420
421 EDFPYTIRLV SEITESNGSS SMATVCGSSL AMMDAGVPLV RPVSGIAMGL ILEQDGFAVL 480
481 SDILGDEDHL GDMDFKVAGT SEGLTSLQMD IKIAGITPAI MEQALAQAKE GRAHILGEMN 540
541 KAMDAPRADV GDFAPKIETI NIPTDKIREV IGSGGKVIRE IVATTGAKVD INDDGVVKVS 600
601 ASDGAKIKAA IDWIKSITDE AEVGKIYDGK VVKVVDFGAF VNFFGAKDGL VHVSQISNER 660
661 VAKPSDVLKE GQMVKVKLLG FDDRGKTKLS MKVVDQETGE DLSKKEAAAE EA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
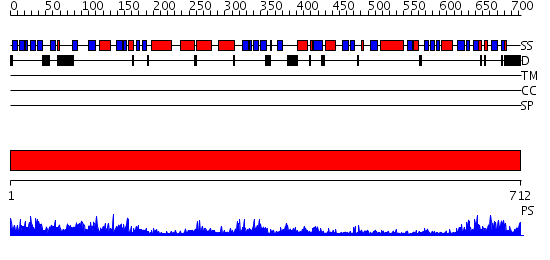
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..712] | 153.0 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)