













| General Information: |
|
| Name(s) found: |
PNP_PSEAE
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Pseudomonas aeruginosa |
| Length: | 701 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPVTKQFQF GQSTVTLETG RIARQATGAV LVTMDDVSVL VTVVGAKSPA EGRDFFPLSV 60
61 HYQEKTYAAG RIPGGFFKRE GRPSEKETLT SRLIDRPIRP LFPEGFMNEV QVVCTVVSTN 120
121 KKSDPDIAAM IGTSAALAIS GIPFAGPIGA ARVGFHPEIG YILNPTYEQL QSSSLDMVVA 180
181 GTEDAVLMVE SEADELTEDQ MLGAVLFAHD EFQAVIRAVK ELAAEAGKPA WDWKAPAENT 240
241 VLVNAIKAEL GEAISQAYTI TIKQDRYNRL GELRDQAVAL FAGEEEGKFP ASEVKDVFGL 300
301 LEYRTVRENI VNGKPRIDGR DTRTVRPLRI EVGVLGKTHG SALFTRGETQ ALVVATLGTA 360
361 RDAQLLDTLE GERKDAFMLH YNFPPFSVGE CGRMGSPGRR EIGHGRLARR GVAAMLPTQD 420
421 EFPYTIRVVS EITESNGSSS MASVCGASLA LMDAGVPVKA PVAGIAMGLV KEGEKFAVLT 480
481 DILGDEDHLG DMDFKVAGTD KGVTALQMDI KINGITEEIM EIALGQALEA RLNILGQMNQ 540
541 VIAKPRAELS ENAPTMLQMK IDSDKIRDVI GKGGATIRGI CEETKASIDI EDDGSVKIYG 600
601 ETKEAAEAAK LRVLAITAEA EIGKIYVGKV ERIVDFGAFV NILPGKDGLV HISQISDKRI 660
661 DKVTDVLQEG QEVKVLVLDV DNRGRIKLSI KDVAAAEASG V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
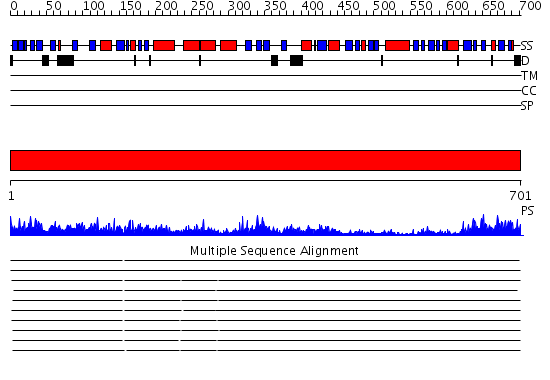
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..701] | 157.0 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)