













| General Information: |
|
| Name(s) found: |
gi|24376022
[NCBI NR]
gi|24375433 [NCBI NR] gi|24351028 [NCBI NR] gi|24350277 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 344 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
transposition, DNA-mediated
[ISS |
| Molecular Function: |
transposase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNYKLISIDL AKTVFQVAAF NPDNTIAFNR KINRPALLDM LRQCEPTLVV MEACYSANYW 60
61 GRTIEKLGHT VKLIPARMVK AMLVGNKNDA NDAVAIGETA GRPKVSLLRP KTVEQQDIQS 120
121 LLRIRERLVE NRTANCNQLR GFLAEYGLIV AKTRRQLMLA IPLILEDADN ELSTVARGFI 180
181 QRLYEFQLYL NEQIDQVAQD ILALAQAMPQ YPLLLTIPGV GPIVASTLLA SINEVNDFKN 240
241 GRQLAAWLGL TPSQYSSGTT NRLGKITKRG NQTLRRLLIH GARTVVNWCD KKDDSLSLWL 300
301 QRLKQRQHVC KVTVALANKM ARMVFVVMSR GQAFDMNKAC KLAT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
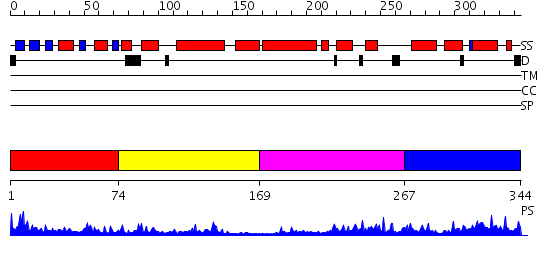
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | 10.510997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [74..168] | 4.585027 | No description for PF01548.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [169..266] | 1.07 | No description for 2eduA was found. | |
| 4 | View Details | [267..344] | 13.565996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)