













| General Information: |
|
| Name(s) found: |
gi|24374961
[NCBI NR]
gi|24349682 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 735 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular response to starvation
[ISS |
| Molecular Function: |
GTP diphosphokinase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSVREAHFN DPDFHLEEWV ARYVSHVDEA QTLLALIAQV DALPAKSPAA KRELLEHARE 60
61 MIEILAPLNM DIETLQAAIL FVVFDAGLLN EEAIKEKFGE PLARLVASVV TMNAIGALKI 120
121 NPNSRSTEPQ IDNIRRMLLA MVEDVRAVVI KLAERVCLLR AVKNADEETR VLLAREIADI 180
181 YAPLANRLGI GQLKWELEDI SFRYLHPDTY KEIAKQLDGK RLDREVFIEN FVEQLQQRLD 240
241 EDHIRAKVYG RPKHIYSIWR KMKGKHLKFD ELFDVRAVRI VTERLQDCYG ALGVVHTLWH 300
301 HIPREFDDYV ANPKPNGYQS IHTVVVGPEG KTVEIQIRTQ DMHEDAELGV AAHWKYKEGN 360
361 HSGKQSGYEE KINWLRKILQ WQEDVVESGN LVEEVRSQVF EDRVYVFTPS GEVVDLPLGS 420
421 TVLDFAYYIH SQVGHKCIGA KVDGRIVPFT YQVETGERIE IITSKHPNPK RDWLNPNLGY 480
481 IRTSRARSKI QHWFKQQDRD KNIIAGKEML EAELARVSLR IKDATIAVER FNMASMDDLL 540
541 AAIGGGDVRL HQVVNHIQSK LRLDEASEED AVEELVKKSQ SKSGTNSRGQ VEVNGVGNLL 600
601 SHIARCCQPV PGDEILGFIT KGRGISVHRS DCEQVKELMR VHPERGVDVV WGENYSGGYR 660
661 MRLRVLAHDR SGLLRDLTSV LAAEKSNVLA MSSSSDIKNQ TAAIELELEL YNLDGLSRVL 720
721 SKLSQVDSVI EARRL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
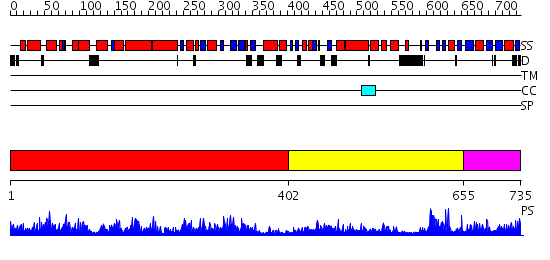
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..401] | 128.0 | Crystal structure of the bifunctional catalytic fragment of RelSeq, the RelA/SpoT homolog from Streptococcus equisimilis. | |
| 2 | View Details | [402..654] | 24.0 | Threonyl-tRNA synthetase (ThrRS), C-terminal domain; Threonyl-tRNA synthetase (ThrRS), N-terminal 'additional' domain; Threonyl-tRNA synthetase (ThrRS), second 'additional' domain; Threonyl-tRNA synthetase (ThrRS) | |
| 3 | View Details | [655..735] | 15.154902 | Crystal structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from E. coli |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)