













| General Information: |
|
| Name(s) found: |
gi|24374902
[NCBI NR]
gi|24349606 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 806 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
proteolysis
[ISS |
| Molecular Function: |
ATP-dependent peptidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTVPTVVPA IPLTSQQIYR RSELQQLSAE CHSTAQLTPL DDIVGQERAQ QAVEFAMGIK 60
61 EKGYNIYAIG QNGLGKRTMM LRYLNRHDPI EPALFDWCYV VNFDDTRSPK VLKLTAGTGL 120
121 EFKKSIEKLM LKLVRALPLA FDNEMYYARA EKLKSQLTQK QEAVLTELSE EAKQKNISLS 180
181 LTMQGDYQMI ALNGDEPHDE ATFSALTEAE RNQFESNING LEVKLRSIIR QNTEWEEEFS 240
241 DTQQEHDEQV AKDVLVHFFK PLKDKYKHQP DVRAFLTGMQ ADILSNLDIF LEESEEQLAL 300
301 AYASLEKKMP RRYQVNVLVS QGEQKYPIVV EESPSYHSLF GYVENATFRG TVFTDFSLIR 360
361 PGSLHKANGG VLLMDAIKVL ERPYVWDGLK RALRSRKLDL SSLEREVSLS GTISLAPEPI 420
421 PLDVKIILFG DHETYQLLQH YDADFIELFR VTADFEDVMD RSDASEAHYA RFISSIVHDN 480
481 NMLHCDRSAI ARIIEYSSRE AQDQQKLSLH SANIANLLRE SNFCAKAVSS KRIEKQHVEQ 540
541 ALKNQQKRVS RLHDNIMESF VNGTTLINTH DKVVGQINAL SVVSTSEHQF GLPNRITATT 600
601 AYGKGEVLDI EHRVKLGGRI HSKGVLILTA YLASVLGKTA QIPLTTYLTF EQSYSGVDGD 660
661 SASMAECCAI ISAISELPLR QDIAITGSMN QFGEAQPIGG VNEKIEGFFD VCQIKGRTSS 720
721 QGVIIPESNI ANLMLRQDIV EAVERGEFHI WAINHVTQAM TLLLGKTAAT VQTNESKHID 780
781 QYAPECIFGI AQQKLNALRA LSKTNS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
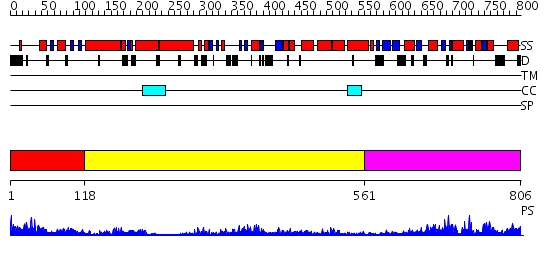
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 18.39794 | Crystal structure of N-terminal domain of E.Coli Lon Protease | |
| 2 | View Details | [118..560] | 46.69897 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [561..806] | 37.0 | Catalytic domain of E.coli Lon protease |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)