













| General Information: |
|
| Name(s) found: |
gi|24374555
[NCBI NR]
gi|24349151 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 498 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
siderophore biosynthetic process
[ISS |
| Molecular Function: |
oxidoreductase activity, acting on single donors with incorporation of molecular oxygen
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTPKKEIDR TIYDLLGIGI GPFNLGLAAL CEPINDLSCL FLDAKTEFDW HPGMLINSSR 60
61 LQTPFMSDLV TMADPTSRFS YLNFAKQTSR LYSFYIRENF FLPRHEYNQY CQWVSKQLSN 120
121 LNFGVKVTQV NYNAGEGIYA VTAIDRRSGK PLTYLCRKLV LGTGTTPYLP DNCPIQDPRV 180
181 MHSASYMQQK TYLQSQSAIT VIGSGQSAAE IFYDLLQDID IYGYQLNWMT RSPRFYPLEY 240
241 TKLTLEMTSP DYVDYFHELS SEKRHRLITE QKSLYKGINA ELINDIYDLL YQKRLISNIQ 300
301 CQLLTNVALT AIDTSGDKLK LQFKHLEQDY ALDQLTSAVV LGTGYQYHLP DFIQGIKSQI 360
361 EFDDHGQLAI QRDYGIDVRG DIFIQNAGLH THGISSPDLG MGCYRNATIL QAVLGYAPYP 420
421 IETRIAFQTF APCNIADAKR QPLESNTHSA VTPSKTRQGL NPSAKSVQQP SIEPQTALRI 480
481 APTGGNVSAL MAPNKEAQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
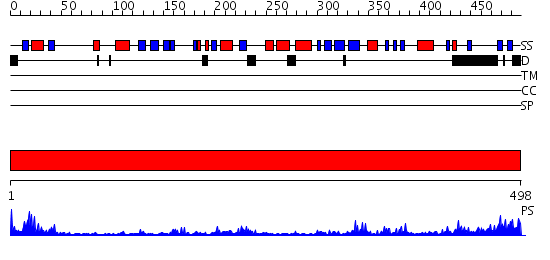
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..498] | 59.69897 | Dihydrolipoamide dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)