













| General Information: |
|
| Name(s) found: |
gi|24374531
[NCBI NR]
gi|24349120 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 557 amino acids |
Gene Ontology: |
|
| Cellular Component: |
provirus
[ISS |
| Biological Process: |
DNA modification
[ISS |
| Molecular Function: |
DNA (cytosine-5-)-methyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRGLIVDNFA GGGGASTGMA WALGRSVDIA INHDQDAIAM HSANHPETLH YCESVFDVDP 60
61 VQATAGKPVA LAWFSPDCKH FSKAKGTKPV NKEIRGLAWV TIRWAMKVRP RVIMLENVEE 120
121 FKTWGPLIQC PVTDAMHPCP ERKGETFNAF VSMLSTGVDA DHPALTECVE TLGLLDSAKL 180
181 IKGLGYKVEF RELRACDYGA PTIRKRLFMI ARCDGQPIVW PQPTHGAPDS EAVKSGKLLP 240
241 WRTAAECIDW SLPCKSIFGR KKPLAENTLR RIAKGIQRFV IDAKEPFIVP NDVQLAPFIT 300
301 EHANASSQRN MPVNEPLRTI CAQVKGGHFA VVQPVLEKVN DNIPTFTEWF AKTKNFGGSY 360
361 EEYVSLYGDD QLTDPVLTAA NICKHYGGNY TGPGDDLNNP LPTVTTVDHN ALITSHMIKL 420
421 RGTNIGFPMD EPAHTITAGG LHLGEVRAFF IKYYGNEQDG VACNEPLHTI TTNDRFGLVM 480
481 IKGEPYQIID IGMRMLEPHE LFACQGFNPE YIINNYNGKS SKKQQVARVG NSVPPQFAEA 540
541 LTRANLPELC TQTAEAA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
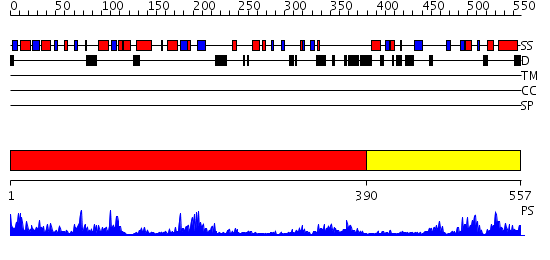
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..389] | 65.154902 | DNA methylase HaeIII | |
| 2 | View Details | [390..557] | 3.18 | DNA methylase HhaI |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)