













| General Information: |
|
| Name(s) found: |
gi|24374516
[NCBI NR]
gi|24349105 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 272 amino acids |
Gene Ontology: |
|
| Cellular Component: |
provirus
[TAS |
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS |
| Molecular Function: |
transcription factor activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNYDDPKKKV GRNIKRLRLK AGIKSQAALA ELCGWKSQSR VGNYEAGTRA VSAIDAETLA 60
61 KVLNVTPAEI LYGENNKDDD FVSMPSAQYS SGPYEEEDPF QLFKPKQELQ NAEWHAGFEL 120
121 WDGDTPLRDD EVALPFYREV ELAAGSGSTF VQENGGCKLR FAKSTLKKSR VEPSNAACVT 180
181 VSGNSMLPVL RHGTTVGVDT SKKSIIDGEM YAIDHDGMLR VKMLYRTPGG GIRIKSYNND 240
241 EFPDEFIPPE KASDIKIIGW VFWWSVLNVW NN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
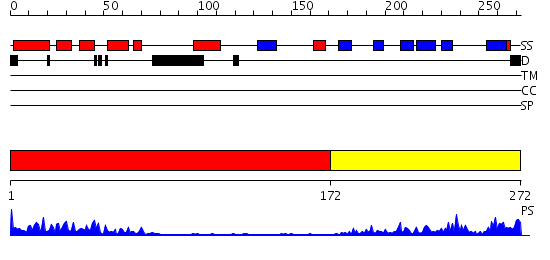
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 11.154902 | Crystal Structure of HTH_3 family Transcriptional Regulator from Vibrio cholerae | |
| 2 | View Details | [172..272] | 13.30103 | No description for 2ho0A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)