













| General Information: |
|
| Name(s) found: |
gi|24374309
[NCBI NR]
gi|24348853 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 291 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
generation of precursor metabolites and energy
[ISS |
| Molecular Function: |
2-hydroxy-3-oxopropionate reductase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKVAFIGLG VMGYPMARHL LNKGHEVTVY NRTFAKAQTW VDTYGGRCCP TPKEAAIGQD 60
61 IVFTCVGNDN DLREVVLGDD GVIHGMALGT VLVDHTTASA DVARELHKVL GEKGIDFLDA 120
121 PVSGGQAGAE NGVLTVMVGG EQAVFERVKP VIEAFARCAE RLGEVGAGQL TKMVNQICIA 180
181 GVVQGLAEAL QFARKAGLDG EKVVEVISKG AAQSWQMENR YKTMWAQNYD FGFAVDWMRK 240
241 DLGIALEEAR RNGSHLPLTA LVDQFYSEVQ AMGGNRWDTS SLLARFEKNT K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
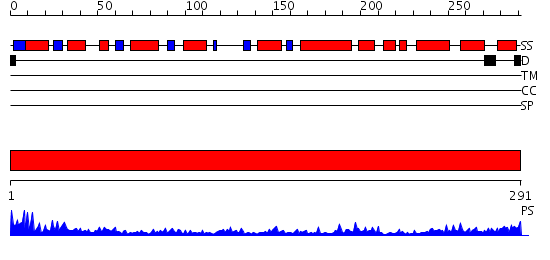
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..291] | 51.0 | X-RAY CRYSTAL STRUCTURE OF TARTRONATE SEMIALDEHYDE REDUCTASE [SALMONELLA TYPHIMURIUM LT2] |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)