













| General Information: |
|
| Name(s) found: |
RUVB_SHEON
[Swiss-Prot]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 334 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Holliday junction helicase complex
[ISS |
| Biological Process: |
DNA recombination
[ISS |
| Molecular Function: |
four-way junction helicase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIEADRLIQP QLQGQDDVID RAMRPKLLDE YTGQDDTRAQ LKVFIQAAKN REEALDHMLI 60
61 YGPPGLGKTT LAMIVANEMG VNIKSTSGPV LEKAGDLAAL LTNLEAGDVL FIDEIHRLSP 120
121 VVEEILYPAM EDYQLDIMIG EGPAARSIKL DLPPFTLVGA TTRAGALTSP LRARFGIPLR 180
181 LEFYNVKDLS TIVTRSAQVM GLAIDSEGAT EIAKRSRGTP RIANRLLRRV RDYAEVQHDG 240
241 AVTQNVAELA LNLLDVDGEG FDYMDRKLLL AIIDKFMGGP VGLDNLAAAI GEERETIEDV 300
301 LEPFLIQQGF IQRTPRGRIA TARAYVHFGM IKPE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
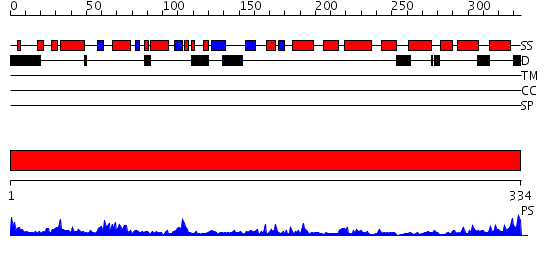
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..334] | 55.0 | Holliday junction helicase RuvB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)