













| General Information: |
|
| Name(s) found: |
gi|24372019
[NCBI NR]
gi|24345883 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 888 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosolic pyruvate dehydrogenase complex
[ISS |
| Biological Process: |
acetyl-CoA biosynthetic process from pyruvate
[ISS |
| Molecular Function: |
pyruvate dehydrogenase (acetyl-transferring) activity
[ISS pyruvate dehydrogenase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEDMLQDVD PLETQEWVDA LQAVLEQEGP ERAHFLLEKL IDKARRNGTH LPYNATTAYL 60
61 NTIPAGQEPH MPGNQEMERR IRAIIRWNAL AMVLRGSKKD LELGGHISSF ASSATIYDVC 120
121 FNHFFRGPNE KDGGDLVYFQ GHIAPGIYAR SFLEGRLSED QLANFRQEVD GKGLSSYPHP 180
181 KLMPDYWQFP TVSMGLGPIQ AIYQARFLKY LTDRGLKDCS AQTVYCFLGD GECDEPEALG 240
241 AIGLAAREEL DNLVFIVNCN LQRLDGPVRG NGKIIQELEG EFRGAGWEVV KVIWGRYWDP 300
301 LLARDTSGKL LQLMEETVDG EYQNCKAKGG AYTREHFFGK YPETAEMVAN MSDDDIWRLN 360
361 RGGHDPVKVY AALDHAQKTK GRPTVILAKT VKGYGLGDAG EGKNIAHNVK KMGIESIRYF 420
421 RDRFNIPIPD DQLEDLPFYH PGPDSEEVKY MMSRRAALHG SVPQRREKFS EELEVPSLKI 480
481 FDSILQGSNG REISSTMAFV RVLTALLKDK KIGKNIVPII PDEARTFGME GLFRQVGIYA 540
541 HEGQKYVPQD SDQVAYYRED KTGQVLQEGI NELGAMSSWV AAATSYSVND TPMIPFYIYY 600
601 SMFGFQRIGD MAWAAGDMRA RGFLVGGTSG RTTLNGEGLQ HQDGHSHVLA NTIPNCITYD 660
661 PTYGYEIAVV VQDGIRRMYG EKQEDIFYYL TTMNENYIQP EMPEGAEEGI VKGIYKLETV 720
721 SGSGKGKVQL LSCGTILEQT RKAAQALAKD FGITADVFSV TSFNELTRDG QAVERWNMLH 780
781 PTQTPKQAYI SQVISSDAPA IAATDYMKIY GEQLRAFMPT DYKVLGTDGF GRSDSRDNLR 840
841 HHFEVDAKFI VIAALKSLVD RKELPVDVLA NAIKEYGIDA DKINPQYA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
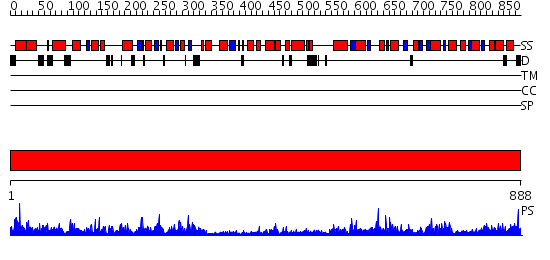
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..888] | 1000.0 | No description for 2qtaA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)