













| General Information: |
|
| Name(s) found: |
gi|24371957
[NCBI NR]
gi|24345803 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 701 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular response to starvation
[ISS |
| Molecular Function: |
guanosine-3',5'-bis(diphosphate) 3'-diphosphatase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYLFEGLKES ASGYLEPEQV ELLKQAYQVA RDAHEGQMRT SGEPYITHPV AVARILAEMR 60
61 LDHETLMAAL LHDTIEDTYV TKEDLAERFG ESVAELVEGV SKLDKLKFRD KKEAQAENFR 120
121 KMMMAMTQDI RVILIKLADR THNMRTLGAL RPDKRRRIAR ETLEIYAPIA NRLGIHNIKT 180
181 ELEDLGFQAY YPMRYRVLKE VVKAARGNRK ELIQGIEAAV LTRLNDAGIK GKVKGREKNL 240
241 YSIYNKMQSK ELQFQEVMDI YAFRVIVDSI DTCYRVLGAM HGLYKPRPGR FKDYIAIPKA 300
301 NGYQSLHTSL FGPHGVPVEI QIRTEDMDQM ADKGVAAHWA YKGGAETGQG TTTQVRARKW 360
361 MQSLLELQQS ASTSFEFVEN VKTELFPEEI YVFTPEGRIL ELPVNATAVD FAYEVHTDVG 420
421 NTCVGARVNR QAYPLSQPLI SGQTVEIITA KGARPNAAWL NFVVTGKARA KIRQVLKSLK 480
481 GDDAIALGRR LLNHALGKTK LDSIPAEQIE KVVRDTKHAS LNALLADIGL GNAMSIVIAQ 540
541 RLIGDNLENQ ESRDSHQMPI RGAEGMLVTF ANCCRPIPGD AVIAHVSQGK GLVVHMENCA 600
601 NIRGYQSEPD KYIPVQWDNV EGVEYQANLR VEIVNHQGAL AKITSIIAAE GSNIHNLSTE 660
661 ERDGRVYLIN LRISVKDRIH LANVMRRIRV LPEVLRTSRN R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
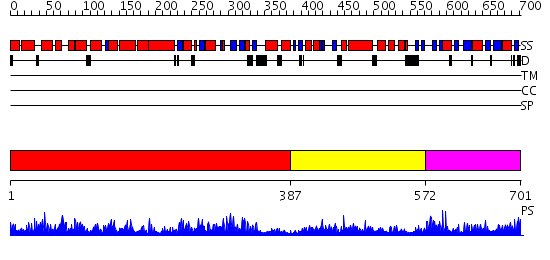
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..386] | 129.0 | Crystal structure of the bifunctional catalytic fragment of RelSeq, the RelA/SpoT homolog from Streptococcus equisimilis. | |
| 2 | View Details | [387..571] | 24.30103 | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with an analogue of threonyl adenylate | |
| 3 | View Details | [572..701] | 12.69897 | Crystal structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from E. coli |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)