













| General Information: |
|
| Name(s) found: |
YRM1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 786 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKRGSLQDR ASPSEETVKK AQKRRKPIKS CAFCRKRKLR CDQQKPMCST CKTRGRSGCL 60
61 YTEKFTHKIE TKELFGSTPN IELLKRIEDL EKRLDDKELT EKDVALSTSP FRNPYANFYY 120
121 LQCKGSGRRI VYGPTSLRTH LSNDDNRFVN TYNQLWSKVK IERNRWKARH KWTMKPETQL 180
181 LEGPPLEKTG SDILQQVCNV LPSFEQSSKI ITDFFNTELE TNEVSEVLDK TKIINDFTSS 240
241 FLPSDELLPN GERRIEKLLP STKKNYYKIG VILMILCIRH FYKNTPEEIE KFLIMLTGLS 300
301 TAKVYFIERA QFLLLKYYHR ELIWACGDDS HMISLVDLLC STAIMLGLHL NIREIYKNQE 360
361 NIVGSMESLE NLWVWIILSD FNVSLNIGRC LAINSSYFQV DECENGERLP KNTNNYSSTV 420
421 FFDQSNSCMG KLKRFLRLAR PMLDQIYDRS AFPDLAENCK KLRNFVETEF HPISYYTDSE 480
481 LISKVPLCEI KVLAQVLNLL LTFYSLRYLI YKEKSVVLEN NILQTILVSF SLVINTTILC 540
541 FNLDEKHFPE FFDHNCVHLP PFMALSLVYT NFLFPRASTG FCAFLYHKLT LFEKGYYLSS 600
601 NIKDQEVTDW DLSTLNIPLD KAMNLLTAFK IHSDIFAKWS NDNNKQLRII MARSYTFVIN 660
661 IALESIYRAV LEKVIKYRTE VENTWLQQLQ DELNGSYSLT DNVNTPIDPS LSDLGVTSAS 720
721 PLAGNSPGLP PEEVRNNSEN ASHNNETGPI ETELAQTISN EFWTAYNLGW EELMSQPDYK 780
781 YLFDTQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
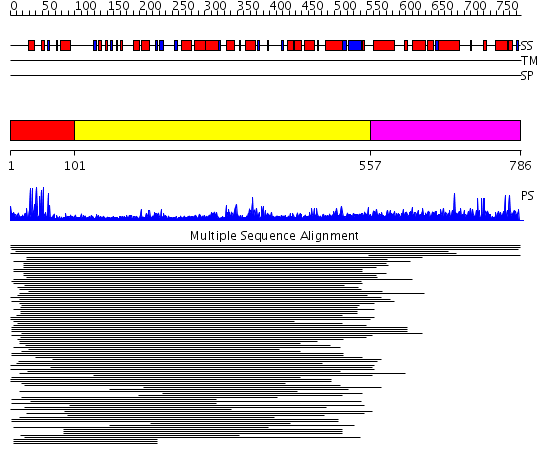
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 70.228787 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [101..556] | 3.144972 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [557..786] | 1.034906 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)