













| General Information: |
|
| Name(s) found: |
UTP10_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1769 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLSDQLAQ VASNNATVAL DRKRRQKLHS ASLIYNSKTA ATQDYDFIFE NASKALEELS 60
61 QIEPKFAIFS RTLFSESSIS LDRNVQTKEE IKDLDNAINA YLLLASSKWY LAPTLHATEW 120
121 LVRRFQIHVK NTEMLLLSTL NYYQTPVFKR ILSIIKLPPL FNCLSNFVRS EKPPTALTMI 180
181 KLFNDMDFLK LYTSYLDQCI KHNATYTNQL LFTTCCFINV VAFNSNNDEK LNQLVPILLE 240
241 ISAKLLASKS KDCQIAAHTI LVVFATALPL KKTIILAAME TILSNLDAKE AKHSALLTIC 300
301 KLFQTLKGQG NVDQLPSKIF KLFDSKFDTV SILTFLDKED KPVCDKFITS YTRSIARYDR 360
361 SKLNIILSLL KKIRLERYEV RLIITDLIYL SEILEDKSQL VELFEYFISI NEDLVLKCLK 420
421 SLGLTGELFE IRLTTSLFTN ADVNTDIVKQ LSDPVETTKK DTASFQTFLD KHSELINTTN 480
481 VSMLTETGER YKKVLSLFTE AIGKGYKASS FLTSFFTTLE SRITFLLRVT ISPAAPTALK 540
541 LISLNNIAKY INSIEKEVNI FTLVPCLICA LRDASIKVRT GVKKILSLIA KRPSTKHYFL 600
601 SDKLYGENVT IPMLNPKDSE AWLSGFLNEY VTENYDISRI LTPKRNEKVF LMFWANQALL 660
661 IPSPYAKTVL LDNLNKSPTY ASSYSSLFEE FISHYLENRS SWEKSCIANK TNFEHFERSL 720
721 VNLVSPKEKQ SFMIDFVLSA LNSDYEQLAN IAAERLISIF ASLNNAQKLK IVQNIVDSSS 780
781 NVESSYDTVG VLQSLPLDSD IFVSILNQNS ISNEMDQTDF SKRRRRRSST SKNAFLKEEV 840
841 SQLAELHLRK LTIILEALDK VRNVGSEKLL FTLLSLLSDL ETLDQDGGLP VLYAQETLIS 900
901 CTLNTITYLK EHGCTELTNV RADILVSAIR NSASPQVQNK LLLVIGSLAT LSSEVILHSV 960
961 MPIFTFMGAH SIRQDDEFTT KVVERTILTV VPALIKNSKG NEKEEMEFLL LSFTTALQHV1020
1021 PRHRRVKLFS TLIKTLDPVK ALGSFLFLIA QQYSSALVNF KIGEARILIE FIKALLVDLH1080
1081 VNEELSGLND LLDIIKLLTS SKSSSEKKKS LESRVLFSNG VLNFSESEFL TFMNNTFEFI1140
1141 NKITEETDQD YYDVRRNLRL KVYSVLLDET SDKKLIRNIR EEFGTLLEGV LFFINSVELT1200
1201 FSCITSQENE EASDSETSLS DHTTEIKEIL FKVLGNVLQI LPVDEFVNAV LPLLSTSTNE1260
1261 DIRYHLTLVI GSKFELEGSE AIPIVNNVMK VLLDRMPLES KSVVISQVIL NTMTALVSKY1320
1321 GKKLEGSILT QALTLATEKV SSDMTEVKIS SLALITNCVQ VLGVKSIAFY PKIVPPSIKL1380
1381 FDASLADSSN PLKEQLQVAI LLLFAGLIKR IPSFLMSNIL DVLHVIYFSR EVDSSIRLSV1440
1441 ISLIIENIDL KEVLKVLFRI WSTEIATSND TVAVSLFLST LESTVENIDK KSATSQSPIF1500
1501 FKLLLSLFEF RSISSFDNNT ISRIEASVHE ISNSYVLKMN DKVFRPLFVI LVRWAFDGEG1560
1561 VTNAGITETE RLLAFFKFFN KLQENLRGII TSYFTYLLEP VDMLLKRFIS KDMENVNLRR1620
1621 LVINSLTSSL KFDRDEYWKS TSRFELISVS LVNQLSNIEN SIGKYLVKAI GALASNNSGV1680
1681 DEHNQILNKL IVEHMKASCS SNEKLWAIRA MKLIYSKIGE SWLVLLPQLV PVIAELLEDD1740
1741 DEEIEREVRT GLVKVVENVL GEPFDRYLD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
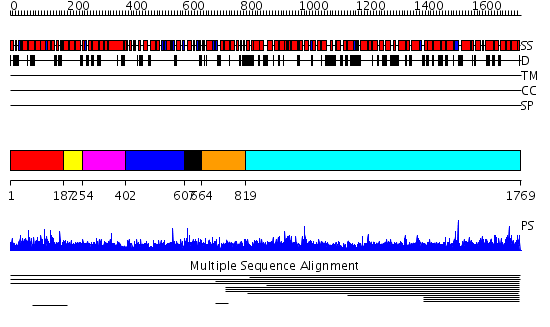
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] | 2.7 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 2 | View Details | [187..253] | 2.7 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [254..401] | 2.7 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [402..606] | 2.7 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [607..663] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [664..818] | 1.005996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [819..1769] | 2.009001 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)