













| General Information: |
|
| Name(s) found: |
UGA3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 528 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNYGVEKLKL KYSKHGCITC KIRKKRCSED KPVCRDCRRL SFPCIYISES VDKQSLKKIK 60
61 ADIQHQLISK KRKHAPDSAQ KAAVATRTRR VGSDEQDNQV YLSKPLEDCI SQKLDSMGLQ 120
121 LYNYYRSHLA NIISIAPMNQ NYYLNIFLPM AHENDGILFA ILAWSANHLS ISSSNELRKD 180
181 EIFVNLANKY TYMSLSHLKT NEGSSACAKL GFLYSLAQIL ILCGSEICQG DVKFWKILLN 240
241 IGKNLIENHV GKDVSRILTT TTEEPSLEER IIFPNFNSVV KYWLIVNFIY HDILNFNTTS 300
301 FPIEQYEKFF QRDQNSLPSS ANFIESIDSP IEEIDPLIGI NKPILLLLGQ VTNLTRFLQT 360
361 MEQEEMLEHG DKILSLQVEI YKLQPSLMAL EHLDDEKKFY YLELFEIMKI STLMFFQLTL 420
421 LKIDKDSLEL QILRNKLDSK LDKVIGTFLE GSLCFPLFIY GVCIQVEDME KKIDLEAKFD 480
481 DILKRYKCYN FQNARLLIRK IWQNEADGIS EHDLVHMIDE LDYNINFA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
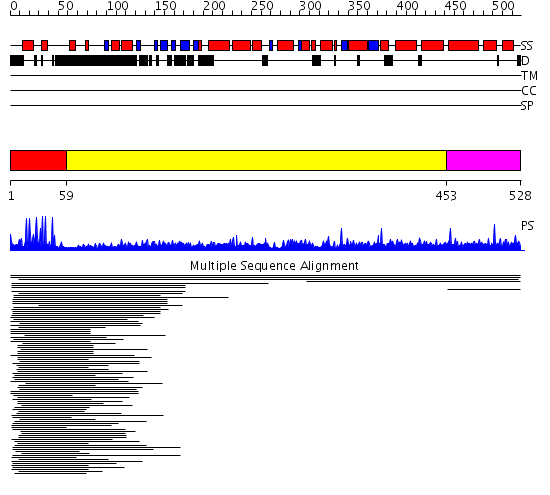
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..58] | 32.201249 | PPR1 | |
| 2 | View Details | [59..452] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [453..528] | 1.004969 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [395..528] | 1.032967 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)