













| General Information: |
|
| Name(s) found: |
UBX2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 584 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPVVNHEDSE FHLSHTEEDK LNEFQVITNF PPEDLPDVVR LLRNHGWQLE PALSRYFDGE 60
61 WKGEPDQMGE PTQTSTPMAE TLVPPALGPR PLLFTASLPV VRPLPANFRN DFRTIGLNGR 120
121 SNTVWSMFES FSYDGNPFLF ILLLIPRIIN RLSATIFTFF CTLLSLHSIS GGGNSGKPKI 180
181 SKVPKAPTRE THIPLAEILG DTKDKDAFCE LKSFKPDISF NEALRIAKEE FKFMLLILVG 240
241 DTYDTDTDTV DVNSKLLLEK ILLNKKTLQY LRKIDNDLII YLKCVHELEP WLVARQLGVR 300
301 NTPEIFLIAN VANKASHSET LPSQRLSILG KLKVNSLNRF LQSLTNVVEK YTPELVVNKT 360
361 EMHELRMSRE IKKLQEDAYK KSLEMDRIKA IEKEKSLKHA QDLKLNSTAR QLKWLKACID 420
421 EIQPFETTGK QATLQFRTSS GKRFVKKFPS MTTLYQIYQS IGCHIYLAVY SSDPAEWSNA 480
481 LQDKIRQLSA DDDMLCFKEG QLETATATTI EELGHIINNE LTSFDLERGK LEFDFELVSP 540
541 FPKYTVHPNE HMSVDQVPQL WPNGSLLVEA LDEEDEEDEE NEEQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
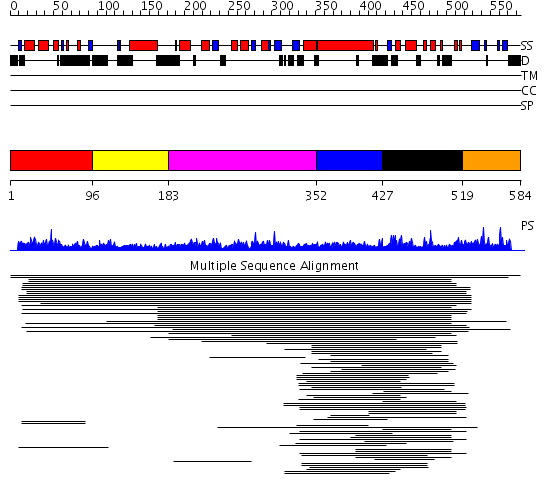
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 2.015997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [96..182] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [183..351] | 1.04498 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [352..426] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [427..518] | 13.154902 | Fas-assosiated factor 1, Faf1 | |
| 6 | View Details | [519..584] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)