













| General Information: |
|
| Name(s) found: |
TBS1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1094 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNMDSGITSS HGSMDKTQKQ SSEWAANQKH NQRVENTRVL MGPAVPAMPP VPSNFPPVPT 60
61 GTIMSPQLSP FPDHRLRHHP LAHMMPADKN FLAYNMESFK SRVTKACDYC RKRKIRCTEI 120
121 EPISGKCRNC IKYNKDCTFH FHEELKRRRE EALNNKGNGK SVKKPRLDKE NKFKDENFDI 180
181 AVRSRNTSST DSSPKLHTNL SQEYIGVSAG KSASDKEDTW PDFVPIDRTV LEKIELNHTK 240
241 VAGKVFVLEE ICKNMKGTIE KLAEKSKIDV IDKEYMKRPK RKQYSKALLT KQKMFHFRQN 300
301 VLSHLTDEEF LSPINEMFTT TFKYSILQTK LVLDFSFRSA SSPSSDNILY PLPRLAIAKR 360
361 LLKNIKCPSL ASLLHIVDVD QCLQFADVHF DPAKGRLTSS QAFLLNICLC LGATVTNFEE 420
421 KQELVDEDNH ETYYFEKFEL WRLRSFTFLN SVYYYHKLSV ARADMTALKA LLLLAKFAQQ 480
481 KISASSAVKV LSVAIKVALD LRLNLHSTYE DLELDEIIKR RRLWCYCFST DKFFSVVLSR 540
541 PPFLKEENTD VLTDESYVEL FRDKILPNLS IKYDDSKLEG VKDIVSVVNL LANHLEYVPY 600
601 IQSYFLSRLS LIESQIYYSC FSIRTTLDDT LDEIIENVLE NQKALDRMRD DLPTILSLEN 660
661 YKENMRILSL DSSKLDFEVS CCTTILLHLR WYHQKITLSL FVISIIGDNL DQRESSRHDI 720
721 AEIIRRSRLD FKRNCIEVLN ILKDFEYYPT VQNEFLYFSL TTVFSMFLYL SEIMVNDEHA 780
781 METGYIIGLL RDTHTRMLGS EERCLSVHNL KWQTSLFFYT FFLRSTMEKF NLTSKYAKFY 840
841 AFDSNYYEGV LNRLVKHTRE SKDDMVELLK TSFINKEKMA AFGSFVTEDQ EKMEVSFNIF 900
901 NEITIQDLNF LQFSSIPKLW ENKTLEPGEE YHHSNGTNTD NNETTGADDT DDNNNNNNNN 960
961 NKNGNNSSST INNNNNNYSN SNNNDNDNNI NDDDDDDDDD DDDDDDDDDD DDNDDDYSNN1020
1021 GADDDEEDDD YDRSLFPTGL ASLLDASYPE RTANDYRDEN EQSNKLFEKI EGHLEHGVFF1080
1081 YDRDFFFKNV CVKM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
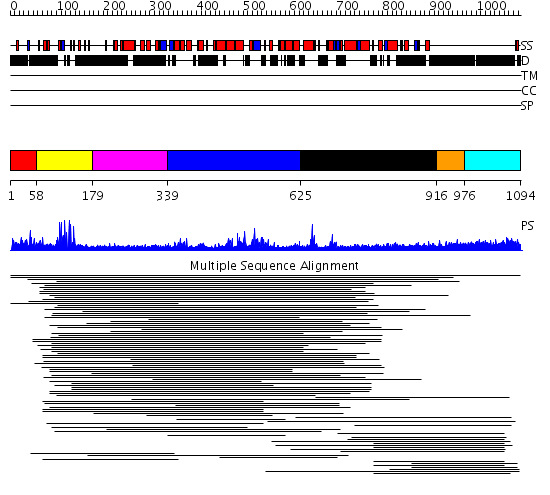
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..57] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [58..178] | 44.512579 | CD2-Lac9 | |
| 3 | View Details | [179..338] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [339..624] | 5.096995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [625..915] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [916..975] | 7.16 | No description for 1l8mA was found. | |
| 7 | View Details | [976..1094] | 7.16 | No description for 1l8mA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)