













| General Information: |
|
| Name(s) found: |
SYC_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNIFIKALRR YTIMSTPKIV QPKWKVPTPQ AKETVLKLYN SLTRSKVEFI PQSGNRGVTW 60
61 YSCGPTVYDA SHMGHARNYV SIDINRRIIQ DYFGYDVQFV QNVTDIDDKI ILRARQNYLF 120
121 DNFVKENDTK FNATVVDKVK TALFQYINKN FTIQGSEIKT IEEFETWLSN ADTETLKLEN 180
181 PKFPMHVTAV QNAIESITKG DSMDAEVAFE KVKDVTVPLL DKELGSTISN PEIFRQLPAY 240
241 WEQKFNDDML SLNVLPPTVT TRVSEYVPEI IDFVQKIIDN GYAYATSDGS VYFDTLKFDK 300
301 SPNHDYAKCQ PWNKGQLDLI NDGEGSLSNF ADNGKKSNND FALWKASKAG EPEWESPWGK 360
361 GRPGWHIECS VMASDILGSN IDIHSGGIDL AFPHHDNELA QSEARFDNQQ WINYFLHTGH 420
421 LHIEGQKMSK SLKNFITIQE ALKKFSPRQL RLAFASVQWN NQLDFKESLI HEVKSFENSM 480
481 NNFFKTIRAL KNDAASAGHI SKKFSPLEKE LLADFVESES KVHSAFCDNL STPVALKTLS 540
541 ELVTKSNTYI TTAGAALKIE PLIAICSYIT KILRIIGFPS RPDNLGWAAQ AGSNDGSLGS 600
601 LEDTVMPYVK CLSTFRDDVR SLAIKKAEPK EFLQLTDKIR NEDLLNLNVA LDDRNGQSAL 660
661 IKFLTNDEKL EIVKLNEEKH ANELAKKQKK LEQQKLREQK ENERKQKAQI KPQDMFKDVT 720
721 LYSAWDEQGL PTKDKDGNDI TKSMTKKLKK QWEQQKKLHE EYFGEDK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
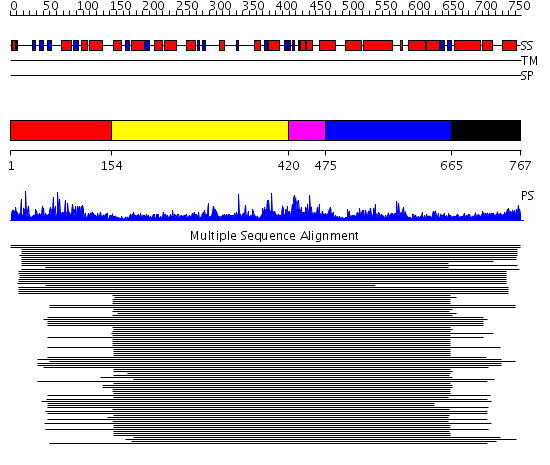
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 2.045757 | No description for 1lleA_ was found. | |
| 2 | View Details | [154..419] | 588.54902 | Cysteinyl-tRNA synthetase (CysRS) | |
| 3 | View Details | [420..474] | 588.54902 | Cysteinyl-tRNA synthetase (CysRS) | |
| 4 | View Details | [475..664] | 588.54902 | Cysteinyl-tRNA synthetase (CysRS) | |
| 5 | View Details | [665..767] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)