













| General Information: |
|
| Name(s) found: |
RRP5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1729 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVASTKRKRD EDFPLSREDS TKQPSTSSLV RNTEEVSFPR GGASALTPLE LKQVANEAAS 60
61 DVLFGNESVK ASEPASRPLK KKKTTKKSTS KDSEASSANS DEARAGLIEH VNFKTLKNGS 120
121 SLLGQISAIT KQDLCITFTD GISGYVNLTH ISEEFTSILE DLDEDMDSDT DAADEKKSKV 180
181 EDAEYESSDD EDEKLDKSNE LPNLRRYFHI GQWLRCSVIK NTSLEPSTKK SKKKRIELTI 240
241 EPSSVNIYAD EDLVKSTSIQ CAVKSIEDHG ATLDVGLPGF TGFIAKKDFG NFEKLLPGAV 300
301 FLGNITKKSD RSIVVNTDFS DKKNKITQIS SIDAIIPGQI VDLLCESITK NGIAGKVFGL 360
361 VSGVVNVSHL RTFSEEDLKH KFVIGSSIRC RIIACLENKS GDKVLILSNL PHILKLEDAL 420
421 RSTEGLDAFP IGYTFESCSI KGRDSEYLYL ALDDDRLGKV HSSRVGEIEN SENLSSRVLG 480
481 YSPVDDIYQL STDPKYLKLK YLRTNDIPIG ELLPSCEITS VSSSGIELKI FNGQFKASVP 540
541 PLHISDTRLV YPERKFKIGS KVKGRVISVN SRGNVHVTLK KSLVNIEDNE LPLVSTYENA 600
601 KNIKEKNEKT LATIQVFKPN GCIISFFGGL SGFLPNSEIS EVFVKRPEEH LRLGQTVIVK 660
661 LLDVDADRRR IIATCKVSNE QAAQQKDTIE NIVPGRTIIT VHVIEKTKDS VIVEIPDVGL 720
721 RGVIYVGHLS DSRIEQNRAQ LKKLRIGTEL TGLVIDKDTR TRVFNMSLKS SLIKDAKKET 780
781 LPLTYDDVKD LNKDVPMHAY IKSISDKGLF VAFNGKFIGL VLPSYAVDSR DIDISKAFYI 840
841 NQSVTVYLLR TDDKNQKFLL SLKAPKVKEE KKKVESNIED PVDSSIKSWD DLSIGSIVKA 900
901 KIKSVKKNQL NVILAANLHG RVDIAEVFDT YEEITDKKQP LSNYKKDDVI KVKIIGNHDV 960
961 KSHKFLPITH KISKASVLEL SMKPSELKSK EVHTKSLEEI NIGQELTGFV NNSSGNHLWL1020
1021 TISPVLKARI SLLDLADNDS NFSENIESVF PLGSALQVKV ASIDREHGFV NAIGKSHVDI1080
1081 NMSTIKVGDE LPGRVLKIAE KYVLLDLGNK VTGISFITDA LNDFSLTLKE AFEDKINNVI1140
1141 PTTVLSVDEQ NKKIELSLRP ATAKTRSIKS HEDLKQGEIV DGIVKNVNDK GIFVYLSRKV1200
1201 EAFVPVSKLS DSYLKEWKKF YKPMQYVLGK VVTCDEDSRI SLTLRESEIN GDLKVLKTYS1260
1261 DIKAGDVFEG TIKSVTDFGV FVKLDNTVNV TGLAHITEIA DKKPEDLSAL FGVGDRVKAI1320
1321 VLKTNPEKKQ ISLSLKASHF SKEAELASTT TTTTTVDQLE KEDEDEVMAD AGFNDSDSES1380
1381 DIGDQNTEVA DRKPETSSDG LSLSAGFDWT ASILDQAQEE EESDQDQEDF TENKKHKHKR1440
1441 RKENVVQDKT IDINTRAPES VADFERLLIG NPNSSVVWMN YMAFQLQLSE IEKARELAER1500
1501 ALKTINFREE AEKLNIWIAM LNLENTFGTE ETLEEVFSRA CQYMDSYTIH TKLLGIYEIS1560
1561 EKFDKAAELF KATAKKFGGE KVSIWVSWGD FLISHNEEQE ARTILGNALK ALPKRNHIEV1620
1621 VRKFAQLEFA KGDPERGRSL FEGLVADAPK RIDLWNVYVD QEVKAKDKKK VEDLFERIIT1680
1681 KKITRKQAKF FFNKWLQFEE SEGDEKTIEY VKAKATEYVA SHESQKADE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
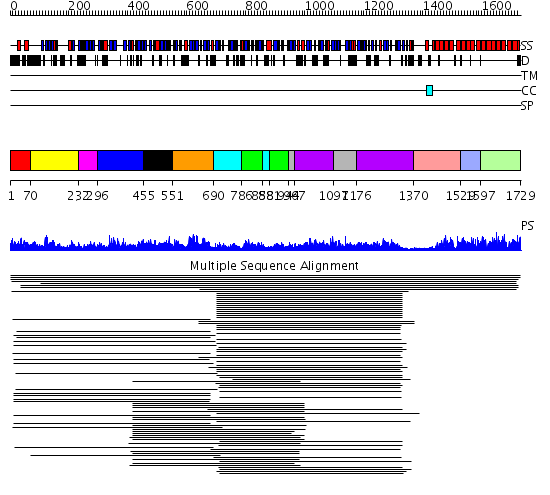
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [70..231] | 2.042991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [232..295] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [296..454] | 1.105996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [455..550] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [551..689] | 56.522879 | S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase | |
| 7 | View Details | [690..785] [858..880] |
75.30103 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 8 | View Details | [786..857] [881..943] |
75.30103 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 9 | View Details | [944..966] [1097..1175] |
75.30103 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 10 | View Details | [967..1096] [1176..1369] |
75.30103 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 11 | View Details | [1370..1528] | 23.69897 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 12 | View Details | [1529..1596] | 23.69897 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 13 | View Details | [1597..1729] | 23.69897 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)