













| General Information: |
|
| Name(s) found: |
PUR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 510 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCGILGIVLA NQTTPVAPEL CDGCIFLQHR GQDAAGIATC GSRGRIYQCK GNGMARDVFT 60
61 QQRVSGLAGS MGIAHLRYPT AGSSANSEAQ PFYVNSPYGI NLAHNGNLVN TASLKRYMDE 120
121 DVHRHINTDS DSELLLNIFA AELEKHNKYR VNNEDVFHAL EGVYRLCRGG YACVGLLAGF 180
181 ALFGFRDPNG IRPLLFGERE NPDGTKDYML ASESVVFKAH NFTKYRDLKP GEAVIIPKNC 240
241 SKGEPEFKQV VPINSYRPDL FEYVYFARPD SVLDGISVYH TRLAMGSKLA ENILKQLKPE 300
301 DIDVVIPVPD TARTCALECA NVLGKPYREG FVKNRYVGRT FIMPNQRERV SSVRRKLNPM 360
361 ESEFKGKKVL IVDDSIVRGT TSKEIVNMAK ESGATKVYFA SAAPAIRYNH IYGIDLTDTK 420
421 NLIAYNRTDE EVAEVIGCER VIYQSLEDLI DCCKTDKITK FEDGVFTGNY VTGVEDGYIQ 480
481 ELEEKRESIA NNSSDMKAEV DIGLYNCADY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
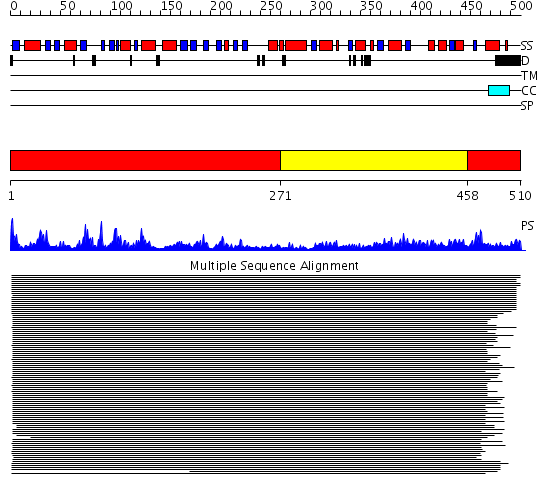
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..270] [458..510] |
1526.0 | Glutamine PRPP amidotransferase, C-terminal domain; Glutamine PRPP amidotransferase, N-terminal domain | |
| 2 | View Details | [271..457] | 1526.0 | Glutamine PRPP amidotransferase, C-terminal domain; Glutamine PRPP amidotransferase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)